Volume 2, Number 3—July 1996
Dispatch
Legionella-Like Amebal Pathogens––Phylogenetic Status and Possible Role in Respiratory Disease
Figure 1
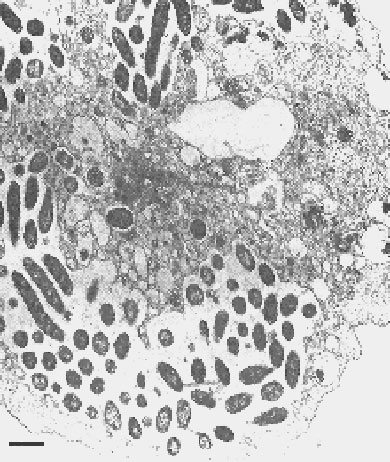
Figure 1. Transmission electron micrograph of Sarcobium lyticum within Acanthamoeba castellanii. Bar = 1 µm. (Reproduced with permission from reference 7.)
References
- Rowbotham TJ. Preliminary report on the pathogenicity of Legionella pneumophila for fresh water and soil amoebae. J Clin Pathol. 1980;33:1179–83. DOIPubMedGoogle Scholar
- Rowbotham TJ. Current views on the relationship between amoebae, legionellae and man. Isr J Med Sci. 1986;22:678–89.PubMedGoogle Scholar
- Chandler FW, Hicklin MD, Blackmon JA. Demonstration of the agent of Legionnaires' disease in tissue. N Engl J Med. 1977;297:1218–20.PubMedGoogle Scholar
- Drozanski W. Fatal bacterial infection in soil amoebae. Acta Microbiol Pol. 1956;5:315–7.PubMedGoogle Scholar
- Drozanski W. Sarcobium lyticum gen. non. sp. nov., an obligate intracellular bacterial parasite of small free living amoebae. Int J Syst Bacteriol. 1991;41:82–7.
- Drozanski W, Chmielewski T. Electron microscope studies of Acanthamoeba castellanii infected with obligate intracellular bacterial parasite. Acta Microbiol Pol. 1979;128:123–33.
- Drozanski W, Madra L, Chmielewski T, Schlecht S, Golecki JR. Evidence for the presence of N-unsubstituted glucosamine residues in the peptidoglycan Sarcobium cytolyticum—an intacellular bacterial parasite of amoebae. Syst Appl Microbiol. 1990;13:220–6.
- Rowbotham TJ. Isolation of Legionella pneumophila from clinical specimens via amoebae, and the interaction of those and other isolates with amoebae. J Clin Pathol. 1983;36:978–86. DOIPubMedGoogle Scholar
- Cianciotto NP, Fields BS. Legionella pneumophila mip gene potentiates intracellular infection of protozoa and human macrophages. Proc Natl Acad Sci U S A. 1992;89:5188–91. DOIPubMedGoogle Scholar
- Fields BS, Barbaree JM, Shotts EB JR, Feeley JC, Morrill WE, Sanden GN, . Comparison of guinea pig and protozoan models for determining virulence of Legionella species. Infect Immun. 1986;53:553–9.PubMedGoogle Scholar
- Cirillo JD, Falkow S, Tompkins LS. Growth of Legionella pneumophila in Acanthamoeba castellanni enhances invasion. Infect Immun. 1994;62:3254–61.PubMedGoogle Scholar
- McDade JE, Shepard CC, Fraser DW, Tsai TR, Redus MA, Dowdle WR, Legionnaires' disease: isolation of a bacterium and demonstration of its role in other respiratory disease. N Engl J Med. 1977;297:1197–203.PubMedGoogle Scholar
- Rowbotham TJ. Legionella-like amoebal pathogens. In: Barbaree JM, Breiman RF, Dufour AP. Legionella: current status and emerging perspectives. Washington, DC: American Society for Microbiology, 1993: 137-40.
- Benson RF, Drozanski WJ, Rowbotham TJ, Fields BS, Bialkowska I, Losos D, Serologic evidence of infection with 9 Legionella-like amoebal pathogens in pneumonia patients. Proceedings of the 95th Annual General Meeting of the American Society for Microbiology, Washington, DC, USA, 1995: Abstract C-200.
- Butler JC, Benson RF, Drozanski WJ, Rowbotham TJ, Bialkowska I, Losos D, Pneumonia associated with Legionella-like amoebal pathogen (LLAP) infection: patient characteristics and clinical manifestations. Proceedings of the 35th Interscience Conference on Antimicrobial Agents and Chemotherapy. American Society for Microbiology, San Francisco, California, USA, 1995: Abstract K-61.
- Garibaldi RA. Epidemiology of community acquired respiratory tract infections in adults: incidence, etiology, and impact. Am J Med. 1985;78:32–7. DOIPubMedGoogle Scholar
- Fass RJ. Aetiology and treatment of community-acquired pneumonia in adults: an historical perspective. J Antimicrob Chemother. 1993;32:17–27.PubMedGoogle Scholar
- Kerttula Y, Leinonen M, Koskela M, Makela PH. The aetiology of pneumonia: application of bacterial serology and basic laboratory methods. J Infect. 1987;14:21–30. DOIPubMedGoogle Scholar
- Springer N, Ludwig W, Drozanski W, Amann R, Schleifer KH.The phylogenetic status of Sarcobium lyticum, an obligate intracellular bacterial parasite of small amoebae. FEMS Microbiol Lett. 1992;96:199–202. DOIGoogle Scholar
- Fry NK, Rowbotham TJ, Saunders NA, Embly TM. Direct amplification and sequencing of the 16S ribosomal DNA of an intacellular Legionella species recovered by amoebal enrichment from the sputum of a patient with pneumonia. FEMS Microbiol Lett. 1991;83:165–8.
- Laussucq S, Schuster D, Alexander WJ, Thacker WL, Wilkinson HW, Spika JS. False positive DNA probe test for Legionella species associated with a cluster of respiratory illnesses. J Clin Microbiol. 1988;26:1442–4.PubMedGoogle Scholar
- Doebbling BN, Bale MJ, Koontz FP, Helms CM, Wenzel RP, Pfaller MA. Prospective evaluation of the Gen-Probe assay for detection of legionellae in respiratory specimens. Eur J Clin Microbiol Infect Dis. 1988;7:748–52. DOIPubMedGoogle Scholar
- Edelstein PH, Bryan RN, Enns RK, Kohne DE, Kacian DL. Retrospective study of Gen-Probe Rapid Diagnostic System for detection of legionellae in frozen clinical respiratory tract samples. J Clin Microbiol. 1987;25:1022–6.PubMedGoogle Scholar
- Fox GE, Wisotzkey JD, Jurtshuk P JR. How close is close; 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992;42:166–70.PubMedGoogle Scholar
- Stackebrandt E, Goebel BM. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–9.
- Giles DL, Fields BS, Newsome AL, Drozanski WJ. Cultivation of Sarcobium lyticum on artificial medium. Proceedings of the 96th Annual General Meeting of the American Society for Microbiology, Washington DC, 1995: Abstract Q-447.
- Smalley DL, Jaquess PA, Layne JS. Selenium-enriched medium for Legionella pneumophila. J Clin Microbiol. 1980;12:32–4.PubMedGoogle Scholar
- Hookey JV, Saunders NA, Fry NK, Birtles RJ, Harrison TJ. Phylogeny of Legionellaceae based on small-subunit ribosomal DNA sequences and proposal of Legionella lytica comb. nov. for Legionella-like amoebal pathogens. Int J Syst Bacteriol. 1996;46:526–31.
- Holt JG, Krieg NR, Sneath PHA, Staley JT, Williams ST. Bergey's manual of determinative bacteriology, Baltimore: Williams & Wilkins, 1994:86.
- Brenner DJ, Feeley JC, Weaver RE. Family VII. Genus I. Legionella Brenner, Steigerwalt and McDade 1979. 658AL. In: Krieg NR, Holt JG editors: Bergey's manual of systematic bacteriology, 1. Baltimore: Williams & Wilkins, 1984:279-88.
- Atschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10.PubMedGoogle Scholar
- Felsenstein J. PHYLIP-Phylogeny inference package. Cladistics. 1989;5:164–6.
- Thacker WL, Plikaytis BB, Wilkinson HW. Identification of 22 Legionella species and 33 serogroups with the slide agglutination test. J Clin Microbiol. 1985;21:779–82.PubMedGoogle Scholar
- Brown SL, Bibb WF, McKinney RM. Retrospective examination of lung tissue specimens for the presence of Legionella organisms: comparison of an indirect fluorescent antibody system with direct fluorescent antibody testing. J Clin Microbiol. 1984;19:468–72.PubMedGoogle Scholar
1Genomic DNA was isolated by standard methods and extracted with phenol/chloroform. A 1400-bp 16S rDNA segment was amplified by using primers specific for Eubacteria and based on a consensus of Legionella 16S rRNA sequences. Each strain was sequenced at least 3 times to ascertain accuracy of obtained results. 16S rRNA sequences for all Legionella species, those of some previously sequenced LLAPs and Coxiella burnetii were obtained from GenBank/EMBL and aligned with sequences generated in this study, using the GCG sequence analysis package (version 8.0). Comparisons were performed at NCBI using the BLAST network service (32). Ambiguous and hypervariable regions were removed before phylogenetic analysis, which was carried out on 53 strains for 1303 nucleotides by the neighbor-joining method from the Phylogeny Inference Package (Phylip) (33), version 3.5. Multiple datasets (X100) were analyzed, and different distance models were compared to ensure reliability. Some of the 16S rRNA sequences included in the data analysis are of unpublished and undescribed stains. Accession numbers for sequences included in this study are as follows: LLAP-1 U64034, LLAP-2 U44909, LLAP-3 X60080, LLAP-4 X97357, LLAP-6 X97357, LLAP-7 U44910, LLAP-8 U64035, LLAP- 9 U44911, LLAP-10 X97363, LLAP-11 X97362, LLAP-12 X97366. L. adelaidensis Z49716, L.anisa X733394, L. birminghamensis Z49717, L. brunensis X73403, L. cherrii X73404, L. cincinnatiensis X73407, Coxiella burnetii M21291, L. donaldsonii Z49724, L.dumoffii X73405, L.erythra Z32638, L. fairfieldensis Z49722, L. feelii X73395, L. geestiana Z49723, L. gormanii Z32639, L. gratiana Z49725, L. hackeliae M36028, L. israelensis X73408, L. jamestowniensis X73409, L. jordanis X73396, L. lansingensis Z49727, L. londiniensis Z49728, L. longbeachae M36029, L. maceachernii X60081, L. micdadei M36032, L. moravica Z49729, L. nautarum Z49730, L. oakridgensis X73397, L. parisiensis Z49731, L. pneumophila M59157, L. quateirensis Z49732, L. worsliensis Z49739, L. quinlivanii Z49733, L. rubrilucens X73398, L. santicrucis Z49735, Sarcobium lyticum X66835, L. shakespearei Z49736, L. spiritensis M36030, L. steigerwaltii X73400, L. sainthelensi Z49734, L. tusconensis Z32644, L. wadsworthii X73401.
2The Remel Poly-ID kit used in this study is designed to identify 22 species and 31 serogroups of Legionella (34,35).