Volume 20, Number 1—January 2014
Research
Genomic Epidemiology of Vibrio cholerae O1 Associated with Floods, Pakistan, 2010
Figure 3
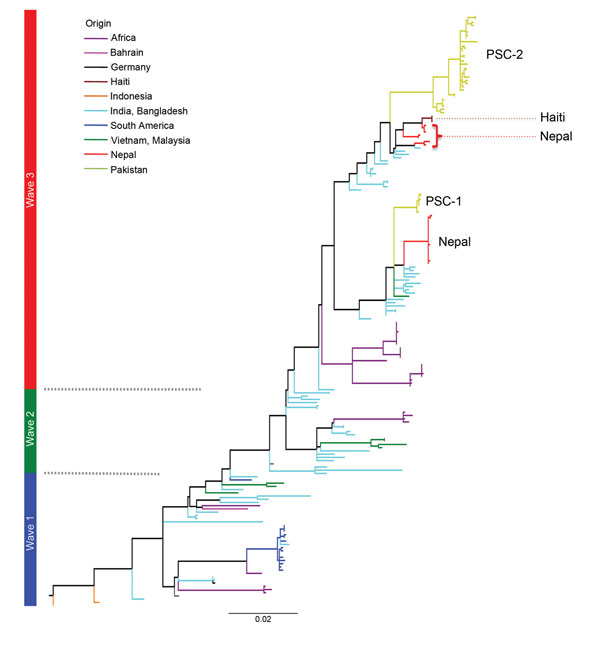
Figure 3. . A single-nucleotide polymorphism–based maximum-likelihood phylogeny showing the position of Vibrio cholerae O1 El Tor from Pakistan in wave 3 of the seventh-pandemic lineage relative to the Haiti and Nepal strains of Hendriksen et al. (23). Waves 1, 2, and 3 are labeled in blue, green, and red respectively. Scale bar indicates substitutions per variable site.
References
- Ahmed K, Shakoori AR. Vibrio cholerae El Tor, Ogawa O1, as the main aetiological agent of two major outbreaks of gastroenteritis in northern Pakistan. J Health Popul Nutr. 2002;20:96–7 .PubMedGoogle Scholar
- Enzensberger R, Besier S, Baumgartner N, Brade V. Mixed diarrhoeal infection caused by Vibrio cholerae and several other enteric pathogens in a 4-year-old child returning to Germany from Pakistan. Scand J Infect Dis. 2005;37:73–5 . DOIPubMedGoogle Scholar
- Jabeen K, Zafar A, Hasan R. Increased isolation of Vibrio cholerae O1 serotype Inaba over serotype Ogawa in Pakistan. East Mediterr Health J. 2008;14:564–70 .PubMedGoogle Scholar
- Singapore Red Cross. Pakistan floods: the deluge of disaster. Facts & figures as of 15 September 2010 [cited 2010 Oct 18]. http://reliefweb.int/report/pakistan/pakistan-floodsthe-deluge-disaster-facts-figures-15-september-2010
- Ramamurthy T, Garg S, Sharma R, Bhattacharya SK, Nair GB, Shimada T, Emergence of novel strain of Vibrio cholerae with epidemic potential in southern and eastern India. Lancet. 1993;341:703–4. DOIPubMedGoogle Scholar
- Mutreja A, Kim DW, Thomson NR, Connor TR, Lee JH, Kariuki S, Evidence for several waves of global transmission in the seventh cholera pandemic. Nature. 2011;477:462–5. DOIPubMedGoogle Scholar
- Waldor MK, Mekalanos JJ. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–4. DOIPubMedGoogle Scholar
- Safa A, Nair GB, Kong RY. Evolution of new variants of Vibrio cholerae O1. Trends Microbiol. 2010;18:46–54. DOIPubMedGoogle Scholar
- Nandi B, Nandy RK, Mukhopadhyay S, Nair GB, Shimada T, Ghose AC. Rapid method for species-specific identification of Vibrio cholerae using primers argeted to the gene of outer membrane protein OmpW. J Clin Microbiol. 2000;38:4145–51 .PubMedGoogle Scholar
- Hoshino K, Yamasaki S, Mukhopadhyay AK, Chakraborty S, Basu A, Bhattacharya SK, Development and evaluation of a multiplex PCR assay for rapid detection of toxigenic Vibrio cholerae O1 and O139. FEMS Immunol Med Microbiol. 1998;20:201–7. DOIPubMedGoogle Scholar
- Winn WC, Allen S, Janda W, Koneman E, Procop G, Schreckenberger P. Curved gram-negative bacilli and oxidase-positive fermenters: Campylobacteraceae and Vibrionaceae. In: Koneman EW, editor. Koneman’s color atlas and textbook of diagnostic microbiology. Washington (DC): Lippincott Williams & Wilkins; 2005. p. 408–28.
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing: nineteenth informational supplement (M100-S19). Wayne (PA): The Institute; 2009.
- Harris SR, Feil EJ, Holden MT, Quail MA, Nickerson EK, Chantratita N, Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327:469–74. DOIPubMedGoogle Scholar
- Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331:430–4. DOIPubMedGoogle Scholar
- Stamatakis A. RAxML-VI-HPC: maximum likelihood–based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–90. DOIPubMedGoogle Scholar
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–91. DOIPubMedGoogle Scholar
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–9. DOIPubMedGoogle Scholar
- Assefa S, Keane TM, Otto TD, Newbold C, Berriman M. ABACAS: algorithm-based automatic contiguation of assembled sequences. Bioinformatics. 2009;25:1968–9. DOIPubMedGoogle Scholar
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10 .PubMedGoogle Scholar
- Carver T, Berriman M, Tivey A, Patel C, Böhme U, Barrell BG, Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics. 2008;24:2672–6 . DOIPubMedGoogle Scholar
- Hendriksen RS, Price LB, Schupp JM, Gillece JD, Kaas RS, Engelthaler DM, Population genetics of Vibrio cholerae from Nepal in 2010: evidence on the origin of the Haitian outbreak. MBio. 2011;2:e00157–11. DOIPubMedGoogle Scholar
- Szabady RL, Yanta JH, Halladin DK, Schofield MJ, Welch RA. TagA is a secreted protease of Vibrio cholerae that specifically cleaves mucin glycoproteins. Microbiology. 2011;157:516–25 . DOIPubMedGoogle Scholar
- Reimer AR, Van Domselaar G, Stroika S, Walker M, Kent H, Tarr C, Comparative genomics of Vibrio cholerae from Haiti, Asia, and Africa. Emerg Infect Dis. 2011;17:2113–21 . DOIPubMedGoogle Scholar
- Pang B, Yan M, Cui Z, Ye X, Diao B, Ren Y, Genetic diversity of toxigenic and nontoxigenic Vibrio cholerae serogroups O1 and O139 revealed by array-based comparative genomic hybridization. J Bacteriol. 2007;189:4837–49. DOIPubMedGoogle Scholar
- Hankins JV, Madsen JA, Giles DK, Brodbelt JS, Trent MS. Amino acid addition to Vibrio cholerae LPS establishes a link between surface remodeling in gram-positive and gram-negative bacteria. Proc Natl Acad Sci U S A. 2012;109:8722–7. DOIPubMedGoogle Scholar
- Saha D, Khan WA, Karim MM, Chowdhury HR, Salam MA, Bennish ML. Single-dose ciprofloxacin versus 12-dose erythromycin for childhood cholera: a randomised controlled trial. Lancet. 2005;366:1085–93. DOIPubMedGoogle Scholar
- Kondo H, Seo N, Yasuda T, Hasizume M, Koido Y, Ninomiya N, Post–flood-infectious diseases in Mozambique. Prehosp Disaster Med. 2002;17:126–33 .PubMedGoogle Scholar
1These authors contributed equally to this article.
Page created: January 03, 2014
Page updated: January 03, 2014
Page reviewed: January 03, 2014
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.