Volume 20, Number 11—November 2014
Dispatch
New Parvovirus in Child with Unexplained Diarrhea, Tunisia
Figure
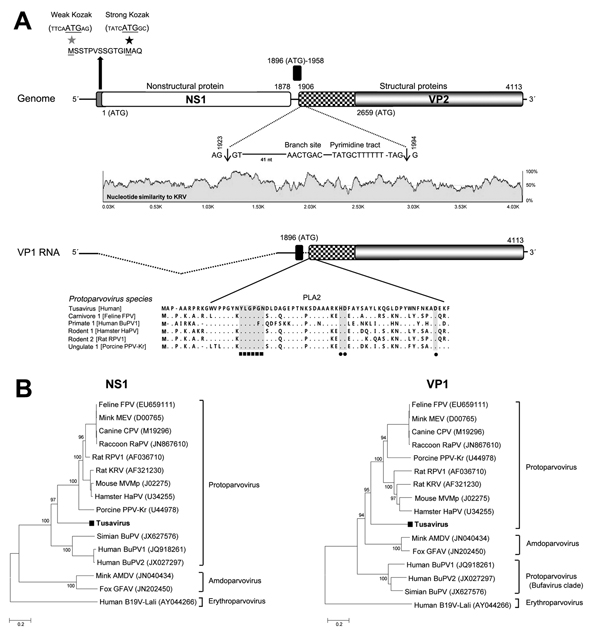
Figure. New parvovirus genome and phylogeny. A) Organization of the Tusavirus genome. Theoretical splicing for expression of viral protein (VP) 1 is shown. The alignment of the PLA2 regions of representatives of 5 protoparvovirus species show the calcium-binding region and catalytic residues in Tusavirus. Pairwise sliding window of percentage nucleotide similarity of Tusavirus aligns with the genetically closest Kilham rat parvovirus. B) Phylogenetic trees generated with nonstructural protein (NS) 1 and VP1 of Tusavirus and of the 5 International Committee on Taxonomy of Viruses–designated species in the Protoparvovirus genus. Scale bar indicatesamino acid substitutions per site. FPV, feline parvovirus; MEV, mink enteritis virus; CPV, canine parvovirus; RaPV, raccoon parvovirus; RPV1, rat parvovirus 1; MVMp, minute virus of mice, prototype; HaPV, hamster parvovirus; PPV-Kr, porcine parvovirus Kresse; Simian BuPV, Simian bufavirus; BuPV1, bufavirus 1; BuPV2, bufavirus 2; AMDV, Aleutian mink disease virus; GFAV, gray fox amdovirus; B19V-Lali, human parvovirus B19-Lali. Bootstrap values (based on 100 replicates) for each node are given if >70%.