Volume 20, Number 12—December 2014
Letter
Ngari Virus in Goats during Rift Valley Fever Outbreak, Mauritania, 2010
Figure
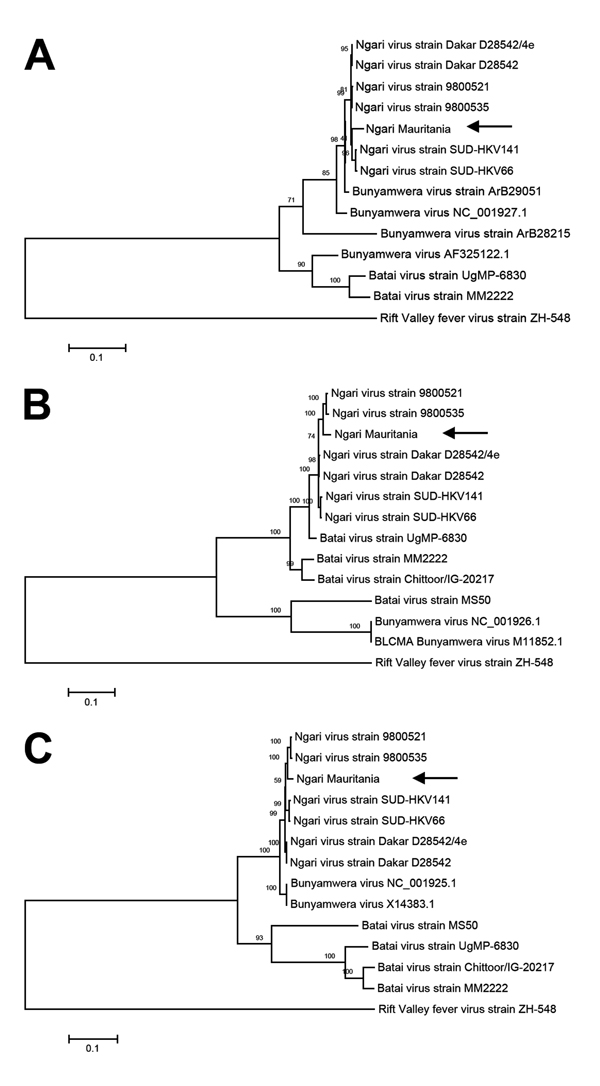
Figure. Phylogenetic tree of Ngari virus–derived A) small (975 bp), B) medium (4,507 bp), and C) large (6,887) segment sequences of Bunyamwera and Batai viruses compared with isolate obtained from a goat in Mauritania in 2010 (arrows). The tree was constructed on the basis of the nucleotide sequences of the 3 complete segments by using the neighbor-joining method (1,000 bootstrap replications). The tree was rooted to the sequence of Rift Valley fever virus strain ZH-548. Scale bars indicate substitutions per nucleotide position.
Page created: November 19, 2014
Page updated: November 19, 2014
Page reviewed: November 19, 2014
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.