Genomic Variability of Monkeypox Virus among Humans, Democratic Republic of the Congo
Jeffrey R. Kugelman
1, Sara C. Johnston
1, Prime M. Mulembakani, Neville Kisalu, Michael S. Lee, Galina Koroleva, Sarah E. McCarthy, Marie C. Gestole, Nathan D. Wolfe, Joseph N. Fair, Bradley S. Schneider, Linda L. Wright, John Huggins, Chris A. Whitehouse, Emile Okitolonda Wemakoy, Jean Jacques Muyembe-Tamfum, Lisa E. Hensley, Gustavo F. Palacios
2
, and Anne W. Rimoin
2
Author affiliations: United States Army Medical Research Institute of Infectious Diseases, Fort Detrick, Maryland, USA (J.R. Kugelman, S.C. Johnston, M.S. Lee, G. Koroleva, S.E. McCarthy, M.C. Gestole, J. Huggins, C.A. Whitehouse, G.F. Palacios); Kinshasa School of Public Health, Kinshasa, Democratic Republic of the Congo (P.M. Mulembakani, E.O. Wemakoy); University of California, Los Angeles, California, USA (N. Kisalu, A.W. Rimoin); Global Viral Forecasting (now known as Metabiota), San Francisco, California, USA (N.D. Wolfe, J.N, Fair, B.S. Schneider); The Eunice Kennedy Shriver National Institute of Child Health and Human Development, Bethesda, Maryland, USA (L.L. Wright); National Institute of Biomedical Research, Kinshasa (J.J. Muyembe-Tamfum); US Food and Drug Administration, Silver Spring, Maryland, USA (L.E. Hensley)
Main Article
Figure 2
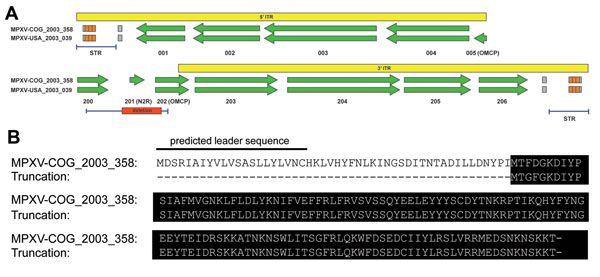
Figure 2. Truncation of OMCPA) Whole-genome deep sequencing revealed a 625-bp deletion directly upstream of the right ITR (red box), which completely removed MPV-Z-N2R (locus 201) and truncated OMCP (MPV-Z-N3R, locus 202)A Western African clade virus, MPXV-USA_2003_039, is shown for comparison with OMCP and N2R copy numberPCR amplification regions for the large deletion and the STRs (right and left) are indicated by the blue bar below each ITR diagramThe yellow box represents the ITR region, and all open reading frames are identified by their MPXV-COG_2003_358 locus number with genes of interest in parenthesisB) Nucleotide alignment of the large deletion with MPXV-COG_2003_358 reference sequenceThe truncated protein created by the deletion is indicated by the black backgroundITR, inverted terminal repeat; MPXV, monkeypox virus; COG, Congo; STR, short terminal repeat; OMCP, orthopoxvirus major histocompatibility complex class I–like protein.
Main Article
Page created: January 17, 2014
Page updated: January 17, 2014
Page reviewed: January 17, 2014
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
