Volume 20, Number 4—April 2014
Research
Rapid Increase in Pertactin-deficient Bordetella pertussis Isolates, Australia
Figure 3
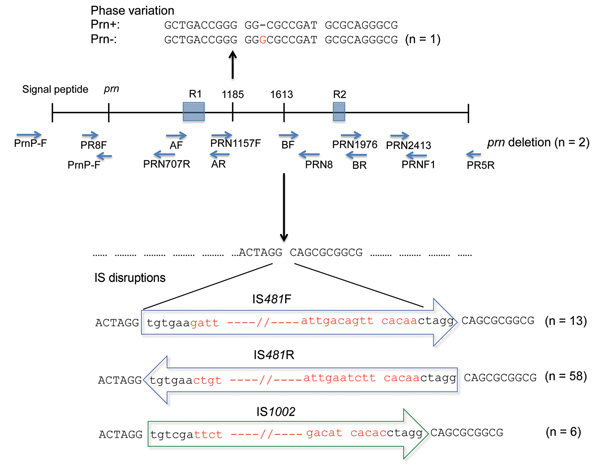
Figure 3. . . Variations in protactin (prn) gene of prn-negative Bordetella pertussis isolates, Australia, 2008–2012, Ninety-six B. pertussis isolates were identified as prn negative. Eighty of these isolates had 1 of 4 mechanisms of prn disruption: IS481 (in forward and reverse directions) and IS1002, which were inserted at the ACTAGG motif within prn, or an extended homopolymeric tract of G residues (n = 1). Lower case letters indicate residues that are conserved in all IS disruptions, and red letters indicate differences in IS disruptions. Positions of nucleotides have been numbered relative to the first start codon of sequence AJ011092 (17). The prn gene of 2 isolates was not amplified by PCR with a combination of primers from published studies (15–19), which indicated a deletion of the entire gene. Sixteen isolates that had no gene disruptions were also observed.
References
- Kurniawan J, Maharjan RP, Chan WF, Reeves PR, Sintchenko V, Gilbert GL, Bordetella pertussis clones identified by multilocus variable-number tandem-repeat analysis. Emerg Infect Dis. 2010;16:297–300. DOIPubMedGoogle Scholar
- Quinn HE, Mahajan D, Hueston L, Campbell P, Menzies RI, Gilbert GL, The seroepidemiology of pertussis in NSW: fluctuating immunity profiles related to changes in vaccination schedules. N S W Public Health Bull. 2011;22:224–9. DOIPubMedGoogle Scholar
- Campbell P, McIntyre P, Quinn H, Hueston L, Gilbert GL, McVernon J. Increased population prevalence of low pertussis toxin antibody levels in young children preceding a record pertussis epidemic in Australia. PLoS ONE. 2012;7:e35874. DOIPubMedGoogle Scholar
- Spokes PJ, Quinn HE, McAnulty JM. Review of the 2008–2009 pertussis epidemic in NSW: notifications and hospitalisations. N S W Public Health Bull. 2010;21:167–73. DOIPubMedGoogle Scholar
- Mooi FR, van der Maas NA, de Melker HE. Pertussis resurgence: waning immunity and pathogen adaptation: two sides of the same coin. Epidemiol Infect. 2013;13:1–10. DOIPubMedGoogle Scholar
- Mooi FR, van Loo IH, van Gent M, He Q, Bart MJ, Heuvelman KJ, Bordetella pertussis strains with increased toxin production associated with pertussis resurgence. Emerg Infect Dis. 2009;15:1206–13. DOIPubMedGoogle Scholar
- Lam C, Octavia S, Bahrame Z, Sintchenko V, Gilbert GL, Lan R. Selection and emergence of pertussis toxin promoter ptxP3 allele in the evolution of Bordetella pertussis. Infect Genet Evol. 2012;12:492–5. DOIPubMedGoogle Scholar
- Octavia S, Maharjan RP, Sintchenko V, Stevenson G, Reeves PR, Gilbert GL, Insight into evolution of Bordetella pertussis from comparative genomic analysis: evidence of vaccine-driven selection. Mol Biol Evol. 2011;28:707–15. DOIPubMedGoogle Scholar
- Octavia S, Sintchenko V, Gilbert GL, Lawrence A, Keil AD, Hogg G, Newly emerging clones of Bordetella pertussis carrying prn2 and ptxP3 alleles implicated in Australian pertussis epidemic in 2008–2010. J Infect Dis. 2012;205:1220–4. DOIPubMedGoogle Scholar
- Hegerle N, Paris AS, Brun D, Dore G, Njamkepo E, Guillot S, Evolution of French Bordetella pertussis and Bordetella parapertussis isolates: increase of Bordetellae not expressing pertactin. Clin Microbiol Infect. 2012;•••:E340–6 PubMed. DOIPubMedGoogle Scholar
- Bouchez V, Brun D, Cantinelli T, Dore G, Njamkepo E, Guiso N. First report and detailed characterization of B. pertussis isolates not expressing pertussis toxin or pertactin. Vaccine. 2009;27:6034–41. DOIPubMedGoogle Scholar
- Otsuka N, Han HJ, Toyoizumi-Ajisaka H, Nakamura Y, Arakawa Y, Shibayama K, Prevalence and genetic characterization of pertactin-deficient Bordetella pertussis in Japan. PLoS ONE. 2012;7:e31985. DOIPubMedGoogle Scholar
- Barkoff AM, Mertsola J, Guillot S, Guiso N, Berbers G, He Q. Appearance of Bordetella pertussis strains not expressing the vaccine antigen pertactin in Finland. Clin Vaccine Immunol. 2012;19:1703–4. DOIPubMedGoogle Scholar
- Weber C, Boursaux-Eude C, Coralie G, Caro V, Guiso N. Polymorphism of Bordetella pertussis isolates circulating for the last 10 years in France, where a single effective whole-cell vaccine has been used for more than 30 years. J Clin Microbiol. 2001;39:4396–403. DOIPubMedGoogle Scholar
- Mooi FR, Hallander H, Wirsing von Konig CH, Hoet B, Guiso N. Epidemiological typing of Bordetella pertussis isolates: recommendations for a standard methodology. Eur J Clin Microbiol Infect Dis. 2000;19:174–81. DOIPubMedGoogle Scholar
- Fry NK, Neal S, Harrison TG, Miller E, Matthews R, George RC. Genotypic variation in the Bordetella pertussis virulence factors pertactin and pertussis toxin in historical and recent clinical isolates in the United Kingdom. Infect Immun. 2001;69:5520–8. DOIPubMedGoogle Scholar
- Mooi FR, van Oirschot H, Heuvelman K, van der Heide HG, Gaastra W, Willems RJ. Polymorphism in the Bordetella pertussis virulence factors P.69/pertactin and pertussis toxin in the Netherlands: temporal trends and evidence for vaccine-driven evolution. Infect Immun. 1998;66:670–5 .PubMedGoogle Scholar
- Boursaux-Eude C, Thiberge S, Carletti G, Guiso N. Intranasal murine model of Bordetella pertussis infection: II. Sequence variation and protection induced by a tricomponent acellular vaccine. Vaccine. 1999;17:2651–60. DOIPubMedGoogle Scholar
- Kinnear SM, Boucher PE, Stibitz S, Carbonetti NH. Analysis of BvgA activation of the pertactin gene promoter in Bordetella pertussis. J Bacteriol. 1999;181:5234–41 .PubMedGoogle Scholar
- Bodilis H, Guiso N. Virulence of pertactin-negative Bordetella pertussis isolates from infants, France. Emerg Infect Dis. 2013;19:471–4 . DOIPubMedGoogle Scholar
- Queenan AM, Cassiday PK, Evangelista A. Pertactin-negative variants of Bordetella pertussis in the United States. N Engl J Med. 2013;368:583–4. DOIPubMedGoogle Scholar
- Pawloski LC, Queenan AM, Cassiday PK, Lynch AS, Harrison M, Shang W, Prevalence and molecular characterization of pertactin-deficient Bordetella pertussis in the US. Clin Vaccine Immunol. 2013. [Epub ahead of print].
- Schmidtke AJ, Boney KO, Martin SW, Skoff TH, Tondella ML, Tatti KM. Population diversity among Bordetella pertussis isolates, United States, 1935–2009. Emerg Infect Dis. 2012;18:1248–55. DOIPubMedGoogle Scholar
- Kurova N, Njamkepo E, Brun D, Tseneva G, Guiso N. Monitoring of Bordetella isolates circulating in Saint Petersburg, Russia between 2001 and 2009. Res Microbiol. 2010;161:810–5. DOIPubMedGoogle Scholar
- Njamkepo E, Cantinelli T, Guigon G, Guiso N. Genomic analysis and comparison of Bordetella pertussis isolates circulating in low and high vaccine coverage areas. Microbes Infect. 2008;10:1582–6. DOIPubMedGoogle Scholar
- Stibitz S. IS481 and IS1002 of Bordetella pertussis create a 6-base-pair duplication upon insertion at a consensus target site. J Bacteriol. 1998;180:4963–6 .PubMedGoogle Scholar
- Parkhill J, Sebaihia M, Preston A, Murphy LD, Thomson N, Harris DE, Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat Genet. 2003;35:32–40. DOIPubMedGoogle Scholar
- Gogol EB, Cummings CA, Burns RC, Relman DA. Phase variation and microevolution at homopolymeric tracts in Bordetella pertussis. BMC Genomics. 2007;8:122. DOIPubMedGoogle Scholar
- Willems R, Paul A, van der Heide HG, ter Avest AR, Mooi FR. Fimbrial phase variation in Bordetella pertussis: a novel mechanism for transcriptional regulation. EMBO J. 1990;9:2803–9 .PubMedGoogle Scholar
- Salaün L, Snyder LA, Saunders NJ. Adaptation by phase variation in pathogenic bacteria. Adv Appl Microbiol. 2003;52:263–301. DOIPubMedGoogle Scholar
- van Gent M, van Loo IH, Heuvelman KJ, de Neeling AJ, Teunis P, Mooi FR. Studies on prn variation in the mouse model and comparison with epidemiological data. PLoS ONE. 2011;6:e18014. DOIPubMedGoogle Scholar
- Bassinet L, Gueirard P, Maitre B, Housset B, Gounon P, Guiso N. Role of adhesins and toxins in invasion of human tracheal epithelial cells by Bordetella pertussis. Infect Immun. 2000;68:1934–41 . DOIPubMedGoogle Scholar