Volume 20, Number 7—July 2014
Dispatch
Cefotaxime-Resistant Salmonella enterica in Travelers Returning from Thailand to Finland
Abstract
During 1993–2011, cefotaxime resistance among Salmonella enterica isolates from patients in Finland increased substantially. Most of these infections originated in Thailand; many were qnr positive and belonged to S. enterica serovar Typhimurium and S. enterica monophasic serovar 4,[5],12:i:-. Although cefotaxime-resistant salmonellae mainly originate in discrete geographic areas, they represent a global threat.
Salmonella spp. are a common cause of foodborne illnesses globally, but illnesses caused by Salmonella infections vary from mild diarrhea (travelers’ diarrhea) to severe generalized infections (1). Certain Salmonella serotypes are more commonly linked to human infections and for example, the monophasic 4,[5],12:i:- variant of S. enterica serovar Typhimurium has caused an increasing number of Salmonella infections in humans during the last decade (2). Antimicrobial agents, usually fluoroquinolones and extended-spectrum cephalosporins, are needed for the treatment of patients with invasive Salmonella infections (3).
The abundant use of antibiotics in human and veterinary medicine and in food production has led to antimicrobial drug resistance (4), and the numbers and proportions of extended-spectrum β-lactamase (ESBL)– and AmpC β-lactamase–producing strains of Enterobacteriaceae have increased worldwide (3,5–7). Although reduced fluoroquinolone susceptibility among S. enterica isolates has increased since the late 1990s (8,9), Salmonella spp. have remained cephalosporin-susceptible. Coexistence of ESBL and plasmid-mediated quinolone resistance genes in Salmonella and in other Enterobacteriaceae genera have been reported and there are existing reports on extended-spectrum cephalosporin-resistant and ESBL-producing Salmonella isolates (3,4,10).
To date, Salmonella isolates that have acquired resistance determinants against fluoroquinolones and extended-spectrum cephalosporins have been reported only anecdotally in Finland. This study describes a systematic analysis of extended-spectrum cephalosporin–resistant Salmonella isolates in Finland during a 19-year period.
During 1993–2011, 43,171 S. enterica isolates were sent to the National Salmonella Reference Centre of the National Institute for Health and Welfare (THL) in Finland. This Salmonella collection contains ≈85% (range 75.9%–91.1%) of all Salmonella isolates collected annually in Finland during the study period. All of these isolates were screened for cefotaxime susceptibility (11). A total of 225 cefotaxime-nonsusceptible S. enterica isolates were identified; 183 of these, collected during 2000–2011, were genotyped. The isolates were screened and serotyped in the Bacteriology Unit at THL.
We confirmed phenotypic ESBL using disk diffusion tests (11). Cefotaxime-nonsusceptible isolates were screened for the ESBL genes TEM, SHV, and CTX-M by PCR (7). CTX-M–positive Escherichia coli, SHV-positive Klebsiella pneumoniae, and TEM-positive E. coli were used as positive ESBL controls. Isolates having only a TEM determinant were further classified by pyrosequencing (12).
We also screened the cefotaxime-nonsusceptible isolates for AmpC production. PCR was used to amplify the AmpC β-lactamase genes CMY, FOX, DHA, ACC, MOX, and EBC by using previously described primers (13). The AmpC multiplex-PCR reaction (50 µL) consisted of 0.2 pmol/µL of each primer, 0.06 U/µL AmpliTaq Gold DNA polymerase, 5 µL AmpliTaq Gold buffer, 2 mM MgCl2, and 0.2 mM dNTP mix (Life Technologies Europe, Espoo, Finland). The PCR program consisted of an initial denaturation at 94°C for 10 minutes, then 38 cycles of DNA denaturation at 94°C for 30 seconds, primer annealing at 64°C for 30 seconds, and extension at 72°C for 1 minute.
We determined susceptibility to the antimicrobial drugs ciprofloxacin, nalidixic acid, and meropenem using the standard agar dilution method according to the Clinical Laboratory and Standards Institute guidelines (11). We screened isolates showing reduced fluoroquinolone susceptibility; specifically, to ciprofloxacin (MIC ≥0.125 µg/mL), that were susceptible or resistant on a low level to nalidixic acid (MIC ≤32 µg/mL) (9) for transferable plasmid-mediated quinolone resistance determinants. We screened the qnrA, qnrB, qnrS, and aac(6′)-Ib-cr genes with a previously described method (14).
We performed the statistical analysis using a log-binomial model and year as an explanatory variable to assess the log-linear trend in time in the percentage/proportion of cefotaxime-nonsusceptible S. enterica isolates. A p value <0.05 was considered significant. Statistical analyses were performed by using IBM SPSS Statistics Version 21 (IBM Corporation, Armonk, NY, USA).
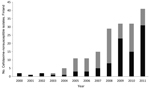
During 1993–2011, we found 225 cefotaxime-nonsusceptible S. enterica isolates and observed a significantly increasing trend (p<0.001) of cefotaxime-nonsusceptible S. enterica isolates (Figure 1). During 1993–1999, 6 S. enterica isolates showed nonsusceptibility to cefotaxime. From the year 2000 onwards, cefotaxime-nonsusceptible isolates were detected more frequently, and in the mid-2000s, the absolute number as well as the proportion of cefotaxime-nonsusceptible Salmonella isolates started to increase rapidly: 55 (2.96%) of 1,858 isolates were positive for this resistance phenotype in 2011 (Figure 1).
During 2000–2011, of the 183 cefotaxime-nonsusceptible isolates, 95 produced ESBL and 88 produced AmpC. The number and proportion of ESBL- and AmpC-positive isolates varied and the number of cefotaxime-nonsusceptible isolates increased (Figure 2). The number of AmpC-positive S. enterica isolates was highest in 2008, and the number of ESBL-positive isolates was highest in 2011. During 2000–2005, 10 ESBL-positive isolates were found; these isolates had been identified in samples collected from travelers from Finland returning from the Mediterranean area, Egypt, and European countries. Isolates positive for the SHV gene mainly originated from Egypt. From 2006 onwards, the main geographic origin of ESBL-positive isolates was Southeast Asia; 61% (52/85) of the ESBL isolates originated from Thailand. During the same time, the CTX-M determinant (72/85 isolates) became more common than SHV. Of the ESBL positive isolates, 44 of 95 belonged to S. enterica ser. Typhimurium or the monophasic 4,[5],12:i:- variant of this serovar; 38 of these originated from Thailand.
AmpC-positive isolates were found from 2003 onwards. During 2003–2004, the AmpC-positive isolates were found in travelers from Finland returning from Spain, India, Mexico, and Africa. From 2005 onwards, the AmpC-positive isolates also commonly originated from Thailand (61/83 isolates). The most common AmpC gene was CMY. Of the AmpC positive isolates, 21 of 88 belonged to S. enterica ser. Typhimurium or a monophasic 4,[5],12:i:- variant of S. enterica ser. Typhimurium serotypes; 8 of these originated from Thailand.
Of the 183 cefotaxime-nonsusceptible Salmonella isolates, 47 had the qnr phenotype; i.e., they showed reduced susceptibility to ciprofloxacin (MIC ≥0.125 µg/mL) but were susceptible or only resistant on a low level to nalidixic acid (MIC ≤32 µg/mL). These isolates were collected from travelers during 2006–2011. Co-resistance to ESBL determinants were detected in 37 isolates: 35 isolates were CTX-M+qnr–positive, including 1 CTX-M+SHV+qnr–positive isolate. Two Salmonella isolates were SHV+qnr positive. Of the 35 CTX-M+qnr–positive isolates, 30 isolates originated from Thailand and 23 of them belonged to the serovar S. enterica ser. Typhimurium or a monophasic 4,[5],12:i:- variant of S. enterica ser. Typhimurium serovars. Ten isolates with an AmpC phenotype were also qnr-positive. Nine of these originated from Southeast Asia and 3 of them were S. enterica ser.Typhimurium or S. enterica ser. 4,[5],12:i:- (Table).
In this study, we described a significant increase (p<0.001) in cefotaxime nonsusceptibility among Salmonella isolates, collected from patients in Finland during 1993–2011. In Salmonella spp., cefotaxime nonsusceptibility is thought to be linked to AmpC-type β-lactamases, and production of ESBLs to be more rare (3). According to our results, ESBL and AmpC production (51.9% vs. 48.1%) were equally common among the cefotaxime-nonsusceptible Salmonella serovars.
During the study period, a change in the geographic origin of cefotaxime-nonsusceptible Salmonella isolates was observed: its predominance in Egypt and the Mediterranean area shifted to Thailand and other Southeast Asian countries. We previously reported that Salmonella isolates with the qnr phenotype are concentrated in Southeast Asia, mainly Thailand (9). In this study, 37 ESBL-positive and 10 AmpC-positive S. enterica isolates were also qnr positive and 40/47 isolates were from Southeast Asia. These results were in concordance with previous reports: ESBL-producing Enterobacteriaceae are commonly isolated from patients returning from Southeast Asia (15) and ESBL and plasmid-mediated quinolone-resistance mechanisms are commonly found in the same plasmids in Enterobacteriaceae and Salmonella (4,6).
We conclude that cefotaxime-nonsusceptible Salmonella isolates are already a threat for travelers to Southeast Asia. Because of the mobile nature of the ESBL and AmpC genes, qnr resistance determinants, and increased travel, this is a worldwide threat, and makes the treatment for invasive Salmonella infections even more challenging.
Dr Gunell is a postdoctoral researcher at the Medical Microbiology and Immunology Unit, University of Turku. Her primary research interests are antimicrobial resistance in Enterobacteriaceae and resistance surveillance.
Acknowledgment
We thank Toni Huovinen, Minna Lamppu, Tuula Randell, and the personnel of the bacteriology unit, University of Turku, for their skillful technical assistance.
References
- Lukinmaa S, Nakari UM, Liimatainen A, Siitonen A. Genomic diversity within phage types of Salmonella enterica ssp. enterica serotypes Enteritidis and Typhimurium. Foodborne Pathog Dis. 2006;3:97–105. DOIPubMedGoogle Scholar
- Soyer Y, Moreno Switt A, Davis MA, Maurer J, McDonough PL, Schoonmaker-Bopp DJ, Salmonella enterica serotype 4,5,12:I:-, an emerging Salmonella serotype that represents multiple distinct clones. J Clin Microbiol. 2009;47:3546–56. DOIPubMedGoogle Scholar
- Sjölund-Karlsson M, Howie R, Krueger A, Rickert R, Pecic G, Lupoli K, CTX-M–-producing non-typhi Salmonella spp. isolated from humans, United States. Emerg Infect Dis. 2011;17:97–9. DOIPubMedGoogle Scholar
- Rodríguez I, Barownick W, Helmuth R, Mendoza MC, Rodicio MR, Schroeter A, Extended-spectrum β-lactamases and AmpC β-lactamases in ceftiofur-resistant Salmonella enterica isolates from food and livestock obtained in Germany during 2003–07. J Antimicrob Chemother. 2009;64:301–9. DOIPubMedGoogle Scholar
- EFSA Panel on Biological Hazards (BIOHAZ). Scientific opinion on the public health risks of bacterial strains producing extended-spectrum β-lactamases and/or AmpC β-lactamases in food and food-producing animals. EFSA Journal. 2011;9:2322 .DOIGoogle Scholar
- Pitout JD, Laupland KB. Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis. 2008;8:159–66. DOIPubMedGoogle Scholar
- Nyberg SD, Österblad M, Hakanen AJ, Huovinen P, Jalava J; The Finnish Study Group for Antimicrobial Resistance. Detection and molecular genetics of extended-spectrum beta-lactamases among cefuroxime-resistant Escherichia coli and Klebsiella spp. isolates from Finland, 2002–2004. Scand J Infect Dis. 2007;39:417–24. DOIPubMedGoogle Scholar
- Hakanen A, Kotilainen P, Huovinen P, Helenius H, Siitonen A. Reduced fluoroquinolone susceptibility in Salmonella enterica serotypes in travelers returning from Southeast Asia. Emerg Infect Dis. 2001;7:996–1003. DOIPubMedGoogle Scholar
- Lindgren MM, Kotilainen P, Huovinen P, Hurme S, Lukinmaa S, Webber MA, Reduced fluoroquinolone susceptibility in Salmonella enterica isolates from travelers, Finland. Emerg Infect Dis. 2009;15:809–12. DOIPubMedGoogle Scholar
- Arlet G, Barrett TJ, Butaye P, Cloeckaert A, Mulvey MR, White DG. Salmonella resistant to extended-spectrum cephalosporins: prevalence and epidemiology. Microbes Infect. 2006;8:1945–54. DOIPubMedGoogle Scholar
- CLSI. Performance standards for antimicrobial susceptibility testing: twentieth informational supplement. CLSI document M100–S20. Wayne (PA): Clinical Laboratory and Standards Institute; 2010.
- Jones CH, Ruzin A, Tuckman M, Visalli MA, Petersen PJ, Bradford PA. Pyrosequencing using the single-nucleotide polymorphism protocol for rapid determination of TEM- and SHV-type extended-spectrum β-lactamases in clinical isolates and identification of the novel β-lactamase genes blaSHV-48, blaSHV-105, and blaTEM-155. Antimicrob Agents Chemother. 2009;53:977–86. DOIPubMedGoogle Scholar
- Pérez-Pérez FJ, Hanson ND. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40:2153–62. DOIPubMedGoogle Scholar
- Gunell M, Webber MA, Kotilainen P, Lilly AJ, Caddick JM, Jalava J, Mechanisms of resistance in nontyphoidal Salmonella enterica strains exhibiting a nonclassical quinolone resistance phenotype. Antimicrob Agents Chemother. 2009;53:3832–6. DOIPubMedGoogle Scholar
- Tham J, Odenholt I, Walder M, Brolund A, Ahl J, Melander E. Extended-spectrum beta-lactamase-producing Escherichia coli in patients with travellers’ diarrhoea. Scand J Infect Dis. 2010;42:275–80. DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleMedline cannot find the journal "EFSA Journal." (in reference 5 "EFSA Panel on Biological Hazards (BIOHAZ), 2011"). Please check the journal name.
Cannot find a title to match the journal "EFSA Journal." (in reference 5 "EFSA Panel on Biological Hazards (BIOHAZ), 2011").
Reference has only first page number. Please provide the last page number if article is longer than one page. (in reference 5 "EFSA Panel on Biological Hazards (BIOHAZ), 2011").
Table of Contents – Volume 20, Number 7—July 2014
| EID Search Options |
|---|
|
|
|
|
|
|