Volume 20, Number 9—September 2014
Dispatch
Novel Circovirus from Mink, China
Figure
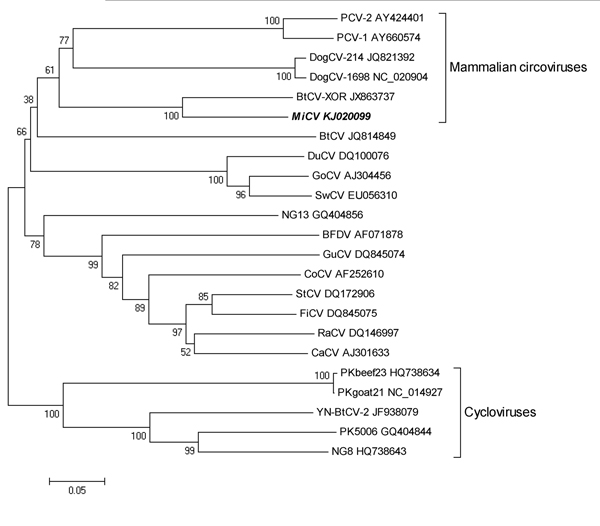
Figure. Phylogenetic tree constructed on the basis of the Rep protein sequence of the mink circovirus by using the neighbor-joining method in MEGA5 (http://www.megasoftware.net). Representative members of the genera Circovirus and Cyclovirus were included in the analysis, and GenBank accession numbers are indicated. Numbers at nodes indicate bootstrap values based on 1,000 replicates. Scale bar indicates nucleotide substitutions per site. The strain sequenced from the mink in Dalian, China, during 2013 (this study) is indicated in boldface italics. PCV-2, porcine circovirus type 2, PCV-1, porcine circovirus type 1, DogCV, dog circovirus; BtCV, bat circovirus; MiCV, mink circovirus; DuCV, duck circovirus; GoCV, goose circovirus; SwCV,swan circovirus; NG13, human stool-associated circular virus NG13; BFDV, beak and feather disease virus; GuCV,gull circovirus; CoCV, columbid circovirus; StCV, starling circovirus; FiCV, finch circovirus; RaCV, raven circovirus; CaCV, canary circovirus; PKbeef23, cyclovirus PKbeef23/PAK/2009; PKgoat21, cyclovirus PKgoat21/PAK/2009; PK 5006, cyclovirus PK5006; NG8, cyclovirus NGchicken8/NGA/2009.
1These authors contributed equally to this article.