Volume 20, Number 9—September 2014
Dispatch
Family Cluster of Middle East Respiratory Syndrome Coronavirus Infections, Tunisia, 2013
Figure 2
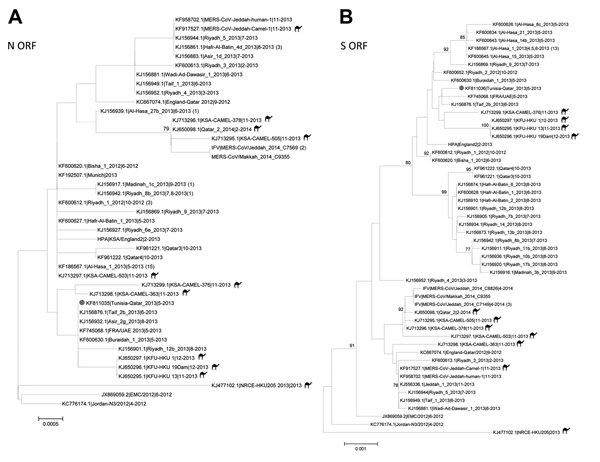
Figure 2. Midpoint-rooted phylogenetic trees of the full-length nucleocapsid (N) (panel A) and spike (S) (panel B) open-reading frames (ORFs) of isolates obtained from index case-patient with Middle East respiratory syndrome coronavirus (MERS-CoV) infection, Tunisia, 2013. Serum and available nucleotide sequences from GenBank and Public Health England (http://www.hpa.org.uk/webw/HPAweb&HPAwebStandard/HPAweb_C/1317136246479) and the Institut Für Virologie (http://www.virology-bonn.de/index.php?id = 46). The estimated neighbor-joining trees were constructed from nucleotide alignments by using MEGA version 6.06 (http://www.megasoftware.net). Sequence names are written as GenBank accession number|virus strain name|month-year of collection. Numbers in parentheses denote the number of additional sequences from viruses isolated from humans that are identical to the listed sequence. Solid circles indicate sequences from MERS-CoV from Tunisia, 2013. Camel icons indicate MERS-CoV sequences derived from isolates from camels. Bootstrap support values (1,000 replicates) ≥75% were plotted at the indicated internal branch nodes. Scale bars indicate number of nucleotide substitutions per site.
1Team members: Philippe Barboza, Alison Bermingham, Julie Fontaine, Alireza Mafi, Babakar Ndoya, and Nahoko Shindo.