Volume 21, Number 1—January 2015
Dispatch
Enzootic Transmission of Yellow Fever Virus, Venezuela
Figure 2
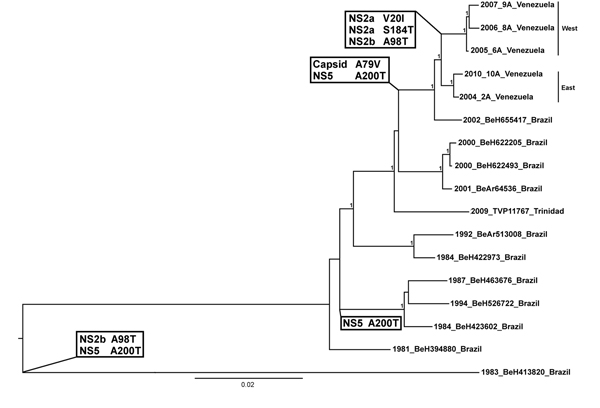
Figure 2. Midpoint rooted Bayesian Markov chain Monte Carlo phylogeny based on yellow fever virus (YFV) complete open reading frame sequences. Numbers at nodes indicate posterior probabilities >0.9. Eastern and western Venezuelan sequences are indicated. Substitutions resulting from nonsynonymous, synapomorphic mutations that define sequences in a clade/lineage are highlighted at relevant nodes. Two substitutions (NS2b A98T and NS5 A200T) occurred in earlier isolates from Brazil. The capsid A79V and NS5 A200T substitutions include the Brazilian isolate BeH655417, which lies directly basal to the Venezuela isolates. This indicates that these substitutions were probably present in the YFV progenitor when it was introduced into Venezuela. Furthermore, substitutions NS2a V10I, NS2a S184T, and NS2b A98T all appear to have arisen after YFV was introduced to Venezuela, further supporting enzootic YFV maintenance there. Taxon/tip labels include year of isolation, strain name and country where the virus was isolated. Scale bar indicates percentage of nucleotide sequence divergence.