Volume 21, Number 12—December 2015
Letter
Isolation of Porcine Epidemic Diarrhea Virus during Outbreaks in South Korea, 2013–2014
Figure
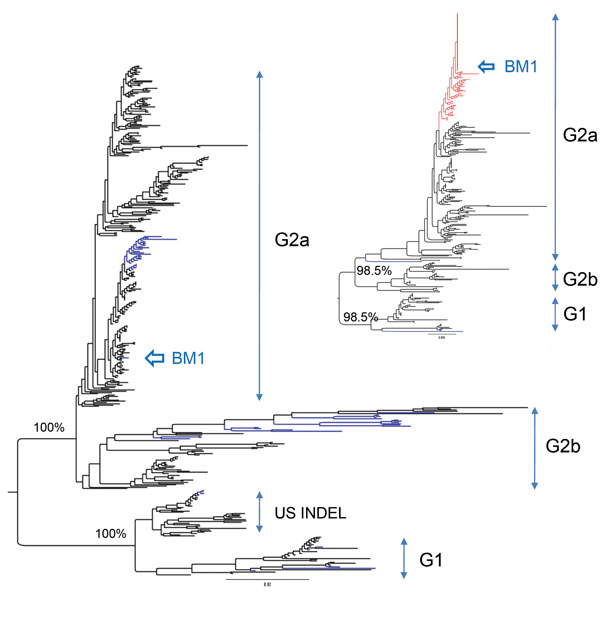
Figure. Maximum-likelihood phylogenetic tree of porcine epidemic diarrhea virus from piglet, South Korea, 2013–2014, constructed on the basis of codon alignment of complete S genes. Inset shows a phylogenetic tree inferred from the complete N genes. Genogroups are shown to the right of each tree. US INDEL is a prototype strain of porcine epidemic diarrhea virus that has insertions and deletions (INDELS) in the spike gene. Scale bars indicate nucleotide substitutions per site.
1These authors contributed equally to this article.
Page created: November 17, 2015
Page updated: November 17, 2015
Page reviewed: November 17, 2015
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.