Severe Ocular Cowpox in a Human, Finland
Paula M. Kinnunen

, Juha M. Holopainen, Heidi Hemmilä, Heli Piiparinen, Tarja Sironen, Tero Kivelä, Jenni Virtanen, Jukka Niemimaa, Simo Nikkari, Asko Järvinen, and Olli Vapalahti
Author affiliations: University of Helsinki, Helsinki, Finland (P.M. Kinnunen, J.M. Holopainen, H. Piiparinen, T. Sironen, T. Kivelä, J. Virtanen, O. Vapalahti); Finnish Defence Forces, Helsinki (P.M. Kinnunen, H. Hemmilä, H. Piiparinen, S. Nikkari); Helsinki University Hospital, Helsinki (J.M. Holopainen, T. Kivelä, A. Järvinen, O. Vapalahti); Natural Resources Institute Finland (Luke), Vantaa, Finland (J. Niemimaa)
Main Article
Figure
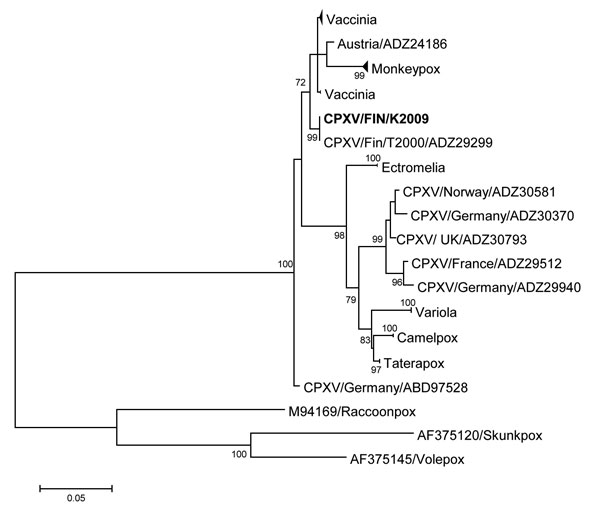
Figure. Phylogenetic tree of orthopoxviruses constructed on the basis of the hemagglutinin gene; boldface indicates the CPXV strain infecting the patient described in this article. The phylogeny shows that the sequence derived from this patient represents a locally circulating strain that shares ancestry with a few other CPXV strains and vaccinia virus. A maximum-likelihood tree was built with 1,000 bootstraps in MEGA 6.06 software (http://www.megasoftware.net/). MEGA was used to estimate the best nucleotide substitution model (general time reversible plus invariable sites). The sequence dataset was compiled from the Virus Pathogen Resource database (http://www.viprbrc.org) and aligned by using MUSCLE (http://www.ebi.ac.uk/Tools/msa/muscle/). Scale bar indicates nucleotide submissions per site. CPXV, cowpox virus.
Main Article
Page created: November 17, 2015
Page updated: November 17, 2015
Page reviewed: November 17, 2015
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
