Volume 21, Number 4—April 2015
Research
Population Structure and Antimicrobial Resistance of Invasive Serotype IV Group B Streptococcus, Toronto, Ontario, Canada
Figure 2
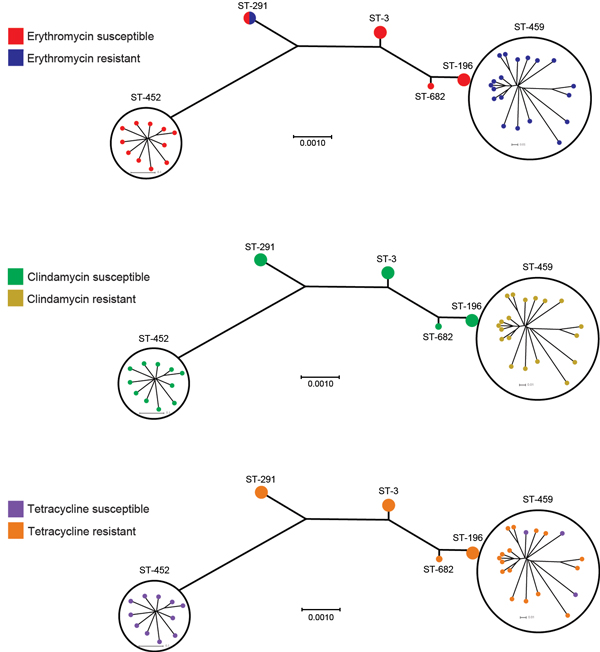
Figure 2. Susceptibility to erythromycin, clindamycin, and tetracycline among serotype IV group B Streptococcus and sequence types (STs), Toronto, Ontario, Canada. All ST-459 strains and 1 ST-291 strain were resistant to erythromycin; we detected ermTR and ermT genes in all erythromycin-resistant strains (Technical Appendix). Resistance to clindamycin was observed only among ST-459 strains. Resistance to tetracycline was common among all STs except ST-452 and correlated with presence of the tetM gene in isolates (Technical Appendix). Scale bars indicate nucleotide substitutions per site.
Page created: April 02, 2015
Page updated: April 02, 2015
Page reviewed: April 02, 2015
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.