Zika Virus Infection, Philippines, 2012
Maria Theresa Alera, Laura Hermann

, Ilya A. Tac-An, Chonticha Klungthong, Wiriya Rutvisuttinunt, Wudtichai Manasatienkij, Daisy Villa, Butsaya Thaisomboonsuk, John Mark Velasco, Piyawan Chinnawirotpisan, Catherine B. Lago, Vito G. Roque, Louis R. Macareo, Anon Srikiatkhachorn, Stefan Fernandez, and In-Kyu Yoon
Author affiliations: Philippines-AFRIMS (Armed Forces Research Institute of Medical Sciences), Cebu City, Philippines (M.T. Alera, J.M. Velasco, C.B. Lago); University of Toronto, Toronto, Ontario, Canada (L. Hermann); AFRIMS, Bangkok, Thailand (L. Hermann, C. Klungthong, W. Rutvisuttinunt, W. Manasatienkij, B. Thaisomboonsuk, P. Chinnawirotpisan, L.R. Macareo, S. Fernandez, I.-K. Yoon); Cebu City Health Department, Cebu City (I.A. Tac-An, D. Villa); Department of Health, Manila, Philippines (V.G. Roque, Jr.); University of Massachusetts Medical School, Worcester, Massachusetts, USA (A. Srikiatkhachorn)
Main Article
Figure
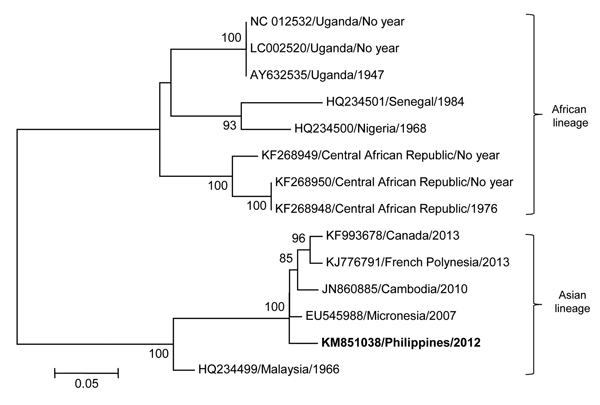
Figure. Maximum-likelihood phylogenetic tree of fragments of Zika virus was determined using the general time-reversible plus gamma distribution plus invariable site model with 13 reference Zika virus strains from GenBank. The contig sequence, obtained from de novo assembly and blastn (http://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome), of the Philippines isolate from 2012 (GenBank accession no. KM851038; bold font) was analyzed against 8 reference strains from Africa (GenBank accession nos. KF268948, KF268949, KF268950, LC002520, AY632535, NC012532, HQ234500, HQ234501) and 5 reference strains from Asia (GenBank accession nos. KJ776791, JN860885, EU545988, HQ234499, KF993678). The year of collection is unknown for several strains from Africa. Bootstrap values >70 are indicated at nodes. Scale bar indicates nucleotide substitutions per site. The drawing is not to scale.
Main Article
Page created: March 17, 2015
Page updated: March 17, 2015
Page reviewed: March 17, 2015
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
