Volume 21, Number 7—July 2015
Letter
Characterization of 3 Megabase-Sized Circular Replicons from Vibrio cholerae
Figure
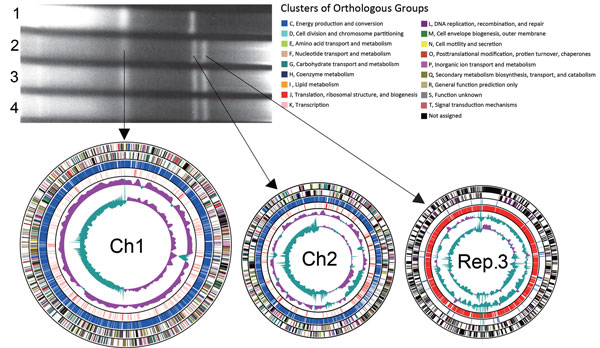
Figure. Pulsed-field gel electrophoresis of intact genomic DNA of Vibrio cholerae isolates and circular representation of the genome of V. cholerae O1 El Tor TSY216, consisting of 3 chromosomes. The preparation of genomic DNA embedded in agarose gels and the protocol for pulsed-field gel electrophoresis have been described previously (5). Arrows indicate DNA bands that correspond to the chromosomes. Lanes: 1, N16961 reference strain carrying Ch1 (2.96 Mb) and Ch2 (1.07 Mb) (2); 2, TSY216; 3, TSY241; 4, TSY421. The first and second outermost 2 circles of each schematic chromosome show the COGs, functional categories of the coding regions of TSY216, in the clockwise and anticlockwise directions, respectively. The third and fourth outermost circles of each schematic chromosome show the coding sequence-assigned (blue) and coding sequences-unassigned (red) functions of the products, respectively. The third and fourth circles show the GC content of the TSY216 sequence and the percent G+C deviation according to the strand, respectively. Ch1, chromosome 1; Ch2, chromosome 2; COGs, clusters of orthologous groups of proteins; Rep.3, novel replicon.
References
- Kolstø AB. Time for a fresh look at the bacterial chromosome. Trends Microbiol. 1999;7:223–6. DOIPubMedGoogle Scholar
- Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–83. DOIPubMedGoogle Scholar
- Yamaichi Y, Iida T, Park KS, Yamamoto K, Honda T. Physical and genetic map of the genome of Vibrio parahaemolyticus: presence of two chromosomes in Vibrio species. Mol Microbiol. 1999;31:1513–21. DOIPubMedGoogle Scholar
- Yamaichi Y, Gerding MA, Davis BM, Waldor MK. Regulatory cross-talk links Vibrio cholerae chromosome II replication and segregation. PLoS Genet. 2011;7:e1002189. DOIPubMedGoogle Scholar
- Okada K, Iida T, Kita-Tsukamoto K, Honda T. Vibrios commonly possess two chromosomes. J Bacteriol. 2005;187:752–7. DOIPubMedGoogle Scholar
- Okada K, Roobthaisong A, Nakagawa I, Hamada S, Chantaroj S. Genotypic and PFGE/MLVA analyses of Vibrio cholerae O1: geographical spread and temporal changes during the 2007–2010 cholera outbreaks in Thailand. PLoS ONE. 2012;7:e30863. DOIPubMedGoogle Scholar
- Reimer AR, Van Domselaar G, Stroika S, Walker M, Kent H, Tarr C, Comparative genomics of Vibrio cholerae from Haiti, Asia, and Africa. Emerg Infect Dis. 2011;17:2113–21. DOIPubMedGoogle Scholar
- Mutreja A, Kim DW, Thomson NR, Connor TR, Lee JH, Kariuki S, Evidence for several waves of global transmission in the seventh cholera pandemic. Nature. 2011;477:462–5. DOIPubMedGoogle Scholar
- Doyle M, Fookes M, Ivens A, Mangan MW, Wain J, Dorman CJ. An H-NS-like stealth protein aids horizontal DNA transmission in bacteria. Science. 2007;315:251–2. DOIPubMedGoogle Scholar
- Chun J, Grim CJ, Hasan NA, Lee JH, Choi SY, Haley BJ, Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc Natl Acad Sci U S A. 2009;106:15442–7. DOIPubMedGoogle Scholar