Identification of Source of Brucella suis Infection in Human by Whole-Genome Sequencing, United States and Tonga
Christine Quance, Suelee Robbe-Austerman

, Tod Stuber, Tom Brignole, Emilio E. DeBess, Laurel Boyd, Brad LeaMaster, Rebekah Tiller, Jenny Draper, Sharon Humphrey, Matthew M. Erdman, and Brad LeaMaster
Author affiliations: National Veterinary Services Laboratories, Ames, Iowa, USA (C. Quance, S. Robbe-Austerman, T. Stuber, M.M. Erdman); United States Department of Agriculture Animal Plant Health Inspection Service, Tumwater, Washington, USA (T. Brignole); Oregon Health Authority Public Health Division, Portland, Oregon, USA (E.E. DeBess, L. Boyd); Oregon Department of Agriculture, Salem, Oregon, USA (B. LeaMaster); Centers for Disease Control and Prevention, Atlanta, Georgia, USA (R. Tiller); Animal Health Laboratory, Ministry for Primary Industries, Wallaceville, New Zealand (J. Draper, S. Humphrey)
Main Article
Figure
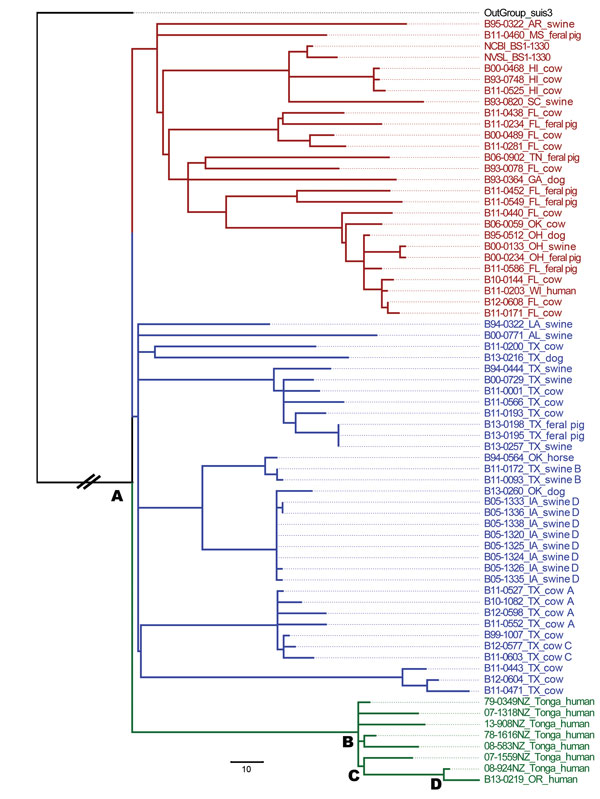
Figure. Maximum-likelihood phylogenetic tree of Brucella suis isolates from the United States and Tonga. The phylogenetic tree was rooted using a truncated B. suis biovar 3 isolate (black text). Red and blue text indicate 59 isolates recovered from US origin sources. Green text indicates the isolate recovered from the immigrant from Tonga residing in Oregon, B13-0219, and 7 additional isolates recovered from patients from Tonga in New Zealand. The first 2 digits of the sample number indicate the year isolated. Isolates recovered from different animals within a herd are labeled with the same letter designation after the species information. The letter A designates the common ancestor between all isolates; B, C, and D identify the common ancestors between the Tonga and Oregon isolates. Scale bar indicates 10 single-nucleotide polymorphisms.
Main Article
Page created: December 18, 2015
Page updated: December 18, 2015
Page reviewed: December 18, 2015
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
