Travel-Associated Vibrio cholerae O1 El Tor, Russia
Konstantin V. Kuleshov

, Sergey O. Vodop’ianov, Vladimir G. Dedkov, Mikhail L. Markelov, Andrey A. Deviatkin, Vladimir D. Kruglikov, Alexey S. Vodop’ianov, Ruslan V. Pisanov, Alexey B. Mazrukho, Svetlana V. Titova, Victor V. Maleev, and German A. Shipulin
Author affiliations: Federal Budget Institute of Science Central Research Institute for Epidemiology, Moscow, Russia (K.V. Kuleshov, V.G. Dedkov, A.A. Deviatkin, V.V. Maleev, G.A. Shipulin); Federal Government Health Institution Rostov-on-Don Plague Control Research Institute, Rostov-on-Don, Russia (S.O. Vodop’ianov, V.D. Kruglikov, A.S. Vodop’ianov, R.V. Pisanov, A.B. Mazrukho, S.V. Titova); Research Institute of Occupational Health, Moscow (M.L. Markelov); Federal Budget Institute Chumakov Institute of Poliomyelitis and Viral Encephalitides, Moscow (A.A. Deviatkin)
Main Article
Figure
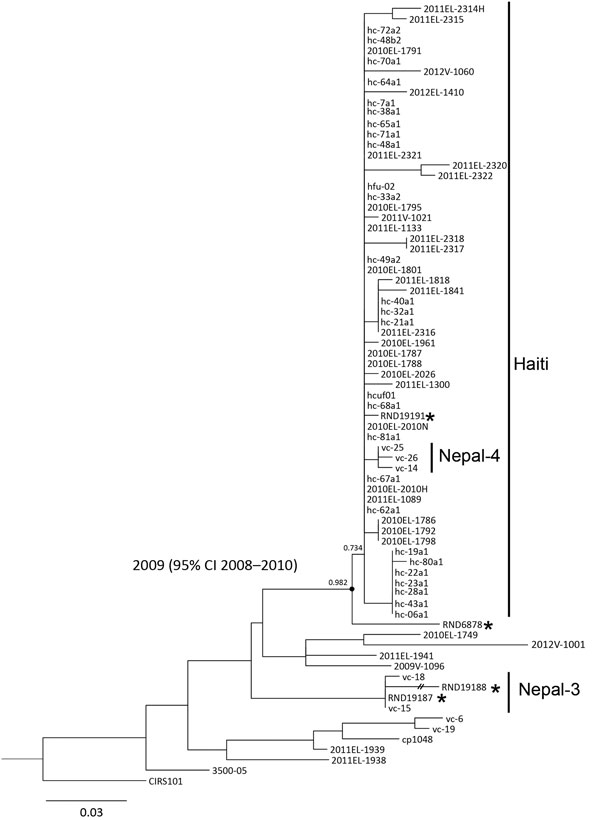
Figure. Maximum-likelihood tree based on an orthologous 193-nt–long high-quality orthologous single-nucleotide polymorphism (hqSNP) matrix of 75 Vibrio cholerae O1 El Tor genomes using the general time-reversible model, withestimation of invariant sites. The phylogenetic tree shows clustering of strains isolated from travel-associated cases of V. cholerae O1 El Tor (asterisks) with isolates collected worldwide. The CIRS101 genome was used as the outgroup. The numbers above nodes represent a statistical branch supports calculated using PhyML (black circle). The internal node between the RND6878 isolate and the Haiti/Nepal-4 clade is labeled with the estimated most recent common ancestor date, which was predicted using BEAST (http://beast.bio.ed.ac.uk). The date range provided represents the 95% CI of the estimate. Scale bar indicates substitutions per variable site.
Main Article
Page created: October 19, 2016
Page updated: October 19, 2016
Page reviewed: October 19, 2016
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
