Volume 22, Number 12—December 2016
Letter
Highly Pathogenic Avian Influenza A(H5N1) Virus among Poultry, Ghana, 2015
Figure
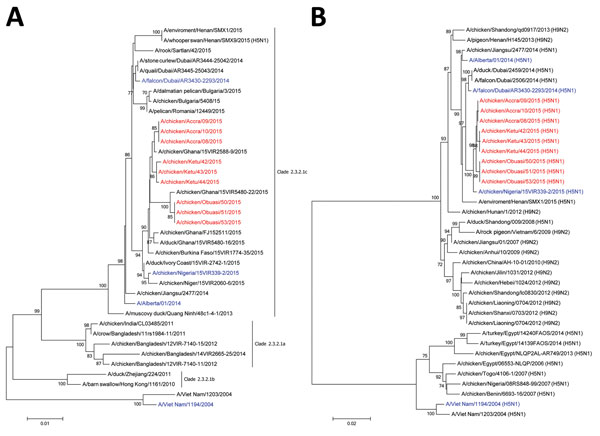
Figure. Phylogenetic analysis of highly pathogenic avian influenza A(H5N1) viruses isolated from poultry in Ghana in 2015: A) hemagglutinin; B) polymerase basic protein 2. Viruses sequenced for this study are in red and reference viruses are in blue; other sequences were downloaded from the Global Initiative on Sharing Avian Influenza Data (http://platform.gisaid.org) and GenBank databases. Evolutionary analyses were conducted with MEGA6 (http://www.megasoftware.net/). Bootstrap values >70% of 500 replicates are shown at the nodes. Scale bars indicate the number of nucleotide substitutions per site.
Page created: November 18, 2016
Page updated: November 18, 2016
Page reviewed: November 18, 2016
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.