Volume 22, Number 12—December 2016
Dispatch
Detection and Genotyping of Coxiella burnetii in Pigs, South Korea, 2014–2015
Cite This Article
Citation for Media
Abstract
We assessed Coxiella burnetii prevalence and genotypes in pigs in South Korea during 2014–2015. Prevalence was low among 1,030 samples tested by ELISA and immunofluorescent assay and 1,124 samples tested by PCR. Despite this finding, possible transmission of C. burnetii from pigs to humans cannot be excluded.
Q fever is a zoonotic disease caused by the extremely infectious bacterium Coxiella burnetii. Humans can be infected by inhalation of infectious aerosols or contaminated dust from infected ruminants or through contact with infected animal products. Ruminants are known as the primary reservoirs for the bacterium. Wildlife may also serve as reservoirs (1). However, epidemiologic data on the occurrence of C. burnetii in pigs are limited. Their susceptibility to C. burnetii infection has been confirmed by the presence of serum antibodies (2), but strong evidence for pigs serving as reservoirs of C. burnetii is lacking. In addition, transmission of C. burnetii from pigs to humans has not been confirmed.
In the veterinary field, commercial immunologic methods are the easiest to interpret and are used at the herd level to detect C. burnetii infection or exposure within a population of animals (3). In South Korea, there have been several studies on C. burnetii in ruminants (4,5), but studies evaluating C. burnetii in pigs are lacking. As first step toward understanding the epidemiology of C. burnetii in pigs, we evaluated the prevalence and genotypes of this bacterium in pigs reared in Gyeongsang Province, South Korea.
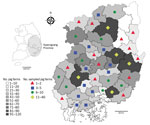
During 2015 in South Korea, a total of 10,332,000 pigs were raised, of which 2,338,521 (22.6%) were raised on 1,134 farms in Gyeongsang Province (6). For this study, we collected 1,030 blood and 97 tissue samples from pigs (645 breeding and 479 fattening pigs) reared on 209 pig farms in Gyeongsang Province during 2014–2015. Sample size was determined using a formula with an expected disease prevalence of 50%, accepted absolute error of 5%, and CI of 99% in a simple random sampling design (Technical Appendix); a minimum of 664 samples were required. Samples were collected by practicing veterinarians during treatment or regular medical checkups; ethical approval was not required. The number of samplings was based on the number of pigs and farms within each of the Province’s administrative districts (Figure 1).
Whole blood was used for PCR; the serum was separated for serologic testing. Lung, lymph node, liver, spleen, and kidney samples were collected for differential diagnosis of diseases in pigs that aborted or had a stillbirth, respiratory symptoms, or weakness.
To detect C. burnetii–positive samples, we used 2 different assays and nested PCR. We used an indirect multispecies ELISA (ID Screen Q Fever Indirect Multi-species Kit; IDvet, Montpellier, France) according to the manufacturer’s instructions to detect C. burnetii antibodies in samples; a sample optical density to positive-control optical density value of >50% was considered positive. We also performed an indirect immunofluorescence assay (IFA), using the Coxiella burnetii (Q Fever) FA Substrate Slide (VMRD, Pullman, WA, USA), as recommended by the manufacturer; titers >64 to phase-1 or phase-2 antigens were considered seropositive. We used the DNeasy Blood and Tissue Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions to extract DNA from whole blood and tissue samples. The Coxiella 16S rRNA gene in extracted DNA was then amplified using nested PCR and sequencing primers (Technical Appendix). We sequenced amplification products with Macrogen (Seoul, South Korea) and analyzed results using sequence alignment programs and statistical methods (Technical Appendix).
Of the 1,030 sampled pigs, 70 (6.8%) were positive for C. burnetii by ELISA (Table); these pigs were from 32 (15.3%) of the 209 sampled farms. Two of the 32 farms had 8 positive pigs each; the other 30 had 1–3 positive pigs each. Fifty-three (5.2%) sampled pigs had samples identified as phase-1 or phase-2 antigen seropositive by IFA; these samples were also seropositive by ELISA. An additional 17 samples seropositive by ELISA were seronegative by IFA. C. burnetii seroprevalence was significantly higher (p<0.0001) in breeding than in fattening pigs by ELISA and IFA.
ELISA and IFA results were in agreement for 1,013 (98.4%) of the 1,030 samples; 53 (5.2%) samples were positive, and 960 (93.2%) were negative. The Cohen κ coefficient was 0.85 (i.e., very good agreement; 95% CI 0.79–0.92).
Three (0.3%) pigs were positive for C. burnetii by PCR; all were breeding pigs and seronegative for C. burnetii. One positive sample was lung tissue from a pig that appeared to have respiratory signs; other respiratory pathogens were also detected in the sample. Acute C. burnetii infection with organ involvement was confirmed by PCR. However, the infection status of seropositive pigs cannot be determined on the basis of a single titer. C. burnetii–seronegative pigs can, however, shed the organism and, thus, might serve as a reservoir for transmission of the bacterium to humans. 16S rRNA gene sequences for the 3 C. burnetii PCR-positive samples (GenBank accession nos. KT945014–16; Figure 2) showed 100% identity with each other; nucleotide sequences showed high (96.6%–96.9%) identity with those of other C. burnetii strains. Phylogenetic analysis showed that the 3 isolates belong to clade A, clustering with previously published C. burnetii sequences (Figure 2).
We found that 6.8%, 5.2%, and 0.3% of tested pig samples in Gyeongsang Province were positive for C. burnetii by ELISA, IFA, and PCR, respectively. These rates of seropositivity are relatively low compared with the rate found in a study in Uruguay, in which 18.4% (83/479) of the blood samples were seropositive by layer microagglutination (7). In that study, the innate susceptibility of pigs to C. burnetii was confirmed during a Q fever epidemic. Seropositivity in our study was, however, higher than that reported in blood tested by IFA in Japan (0/396 samples) (8) and by complement fixation in Bulgaria (0.05%; 1/1,809 samples) (9).
In C. burnetii–positive animals, bacterial burden is highest in birth products. We did not test such tissues; however, the positivity rate in our study was similar to that (0/16) in a previous examination of pig placentas by real-time PCR in the Netherlands (10). In our study, the PCR-positive pig samples did not test positive by serologic methods.
Similar to results from a previous study (11), our results showed that IFA was less sensitive than ELISA at detecting C. burnetii in serum. However, serologic diagnosis of coxiellosis in animals is complicated. Animals can maintain seropositivity after acute infection has cleared, and they can seroconvert without shedding (12); thus, serologic methods are not useful for determining which animals currently pose a risk for transmission.
In our study, seroprevalence among breeding pigs was significantly high (p<0.05). In addition, only breeding pigs were positive for C. burnetii by PCR. Because of pregnancy stress, breeding pigs probably experienced a recrudescent infection, making them more likely to shed the organism. A study on the epidemiology of Q fever suggested that breeding pigs can cause infection in humans (13).
The genus Coxiella is divided into 4 highly divergent genetic clades (A–D); C. burnetii belongs to clade A (14). Phylogenetic analysis showed that the 3 C. burnetii isolates in our study were closely related to clade A strains from the United States, Japan, and Greenland, indicating a close epidemiologic link.
Although the number of C. burnetii–positive pigs was low in our study, a previous study identified contact with pigs as a risk factor for C. burnetii seropositivity in humans (15). Therefore, pigs may serve as potential reservoirs for C. burnetii. However, several questions remain unanswered regarding the epidemiology of C. burnetii infection in pigs and possible transmission to humans. Additional investigations of the infection prevalence in other animals are necessary to understand the epidemiology of C. burnetii.
Dr. Seo works as a research scientist at the Animal and Plant Quarantine Agency, Gimcheon, South Korea. His primary research interests are vectorborne diseases.
Acknowledgment
This research was supported by a grant from the Basic Science Research Program through the National Research Foundation of Korea (NRF), which is funded by the Ministry of Education (grant no. NRF-2016R1D1A1B02015366).
References
- McQuiston JH, Childs JE. Q fever in humans and animals in the United States. Vector Borne Zoonotic Dis. 2002;2:179–91.DOIPubMedGoogle Scholar
- Marmion BP, Stoker MG. The epidemiology of Q fever in Great Britain; an analysis of the findings and some conclusions. Br Med J. 1958;2:809–16.DOIPubMedGoogle Scholar
- Guatteo R, Beaudeau F, Joly A, Seegers H. Coxiella burnetii shedding by dairy cows. Vet Res. 2007;38:849–60.DOIPubMedGoogle Scholar
- Ouh IO, Seo MG, Do JC, Kim IK, Cho MH, Kwak DM. Seroprevalence of Coxiella burnetii in bulk-tank milk and dairy cattle in Gyeongbuk Province, Korea. Korean Journal of Veterinary Service. 2013;36:243–8 .DOIGoogle Scholar
- Jung BY, Seo MG, Lee SH, Byun JW, Oem JK, Kwak D. Molecular and serologic detection of Coxiella burnetii in native Korean goats (Capra hircus coreanae). Vet Microbiol. 2014;173:152–5.DOIPubMedGoogle Scholar
- Korean Statistical Information Service. Statistics Korea. Number of pig and pig farms by city and province in the third quarter of 2015 [in Korean]. 2015 [cited 2016 Feb 20]. http://kosis.kr/statHtml/statHtml.do?orgId=101&tblId=DT_1EO051&conn_path=I2&language=en/
- Somma-Moreira RE, Caffarena RM, Somma S, Pérez G, Monteiro M. Analysis of Q fever in Uruguay. Rev Infect Dis. 1987;9:386–7.DOIPubMedGoogle Scholar
- Htwe KK, Amano K, Sugiyama Y, Yagami K, Minamoto N, Hashimoto A, et al. Seroepidemiology of Coxiella burnetii in domestic and companion animals in Japan. Vet Rec. 1992;131:490.DOIPubMedGoogle Scholar
- Martinov SP, Pandarov S, Popov GV. Seroepizootology of Q fever in Bulgaria during the last five years. Eur J Epidemiol. 1989;5:425–7.DOIPubMedGoogle Scholar
- Roest HI, van Solt CB, Tilburg JJ, Klaassen CH, Hovius EK, Roest FT, et al. Search for possible additional reservoirs for human Q fever, The Netherlands. Emerg Infect Dis. 2013;19:834–5.DOIPubMedGoogle Scholar
- Péter O, Dupuis G, Burgdorfer W, Peacock M. Evaluation of the complement fixation and indirect immunofluorescence tests in the early diagnosis of primary Q fever. Eur J Clin Microbiol. 1985;4:394–6.DOIPubMedGoogle Scholar
- McQuiston JH, Childs JE, Thompson HA. Q fever. J Am Vet Med Assoc. 2002;221:796–9.DOIPubMedGoogle Scholar
- Stoker MG, Thompson JF. An explosive outbreak of Q fever. Lancet. 1953;1:137–9.DOIPubMedGoogle Scholar
- Duron O, Noël V, McCoy KD, Bonazzi M, Sidi-Boumedine K, Morel O, et al. The recent evolution of a maternally-inherited endosymbiont of ticks led to the emergence of the Q fever pathogen, Coxiella burnetii. PLoS Pathog. 2015;11:e1004892.DOIPubMedGoogle Scholar
- Whitney EA, Massung RF, Candee AJ, Ailes EC, Myers LM, Patterson NE, et al. Seroepidemiologic and occupational risk survey for Coxiella burnetii antibodies among US veterinarians. Clin Infect Dis. 2009;48:550–7.DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleTable of Contents – Volume 22, Number 12—December 2016
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Dongmi Kwak, College of Veterinary Medicine, Kyungpook National University, 80 Daehakro, Bukgu, Daegu 41566, South Korea
Top