Volume 22, Number 2—February 2016
Letter
Detection of Influenza D Virus among Swine and Cattle, Italy
Figure
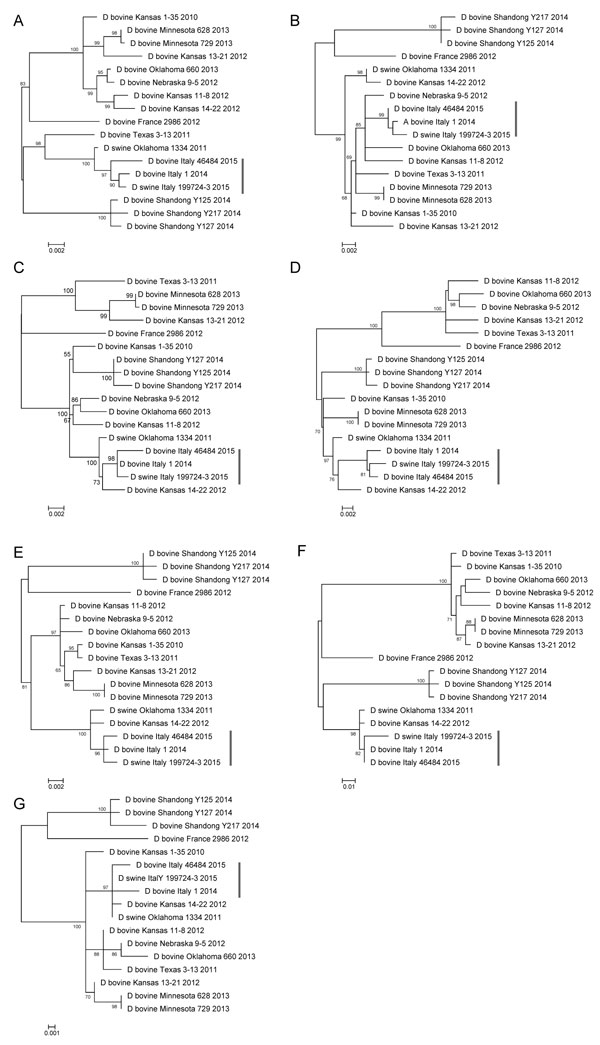
Figure. Phylogenetic trees of the 7 genes of influenza D viruses obtained from 1 sow and 2 cattle in Italy (vertical bars) and comparison isolates retrieved from GenBank. A) Polymerase basic (PB) 2: 2,319 nt; B) PB1: 1,434 nt; C) P3: 2,133 nt; D) glycoprotein hemagglutininesterase: 1,995 nt; E) nucleoprotein: 1,659 nt; F) polymerase 42: 1,164 nt; G) nonstructural: 732 nt. Genes were trimmed and aligned, then phylogenetically analyzed by using the maximum-likelihood method. Sequences are listed by their host, country, strain name, and collection year. Scale bars indicate nucleotide substitutions per site.
1These authors contributed equally to this article.
Page created: January 19, 2016
Page updated: January 19, 2016
Page reviewed: January 19, 2016
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.