Volume 22, Number 2—February 2016
Letter
AP92-like Crimean-Congo Hemorrhagic Fever Virus in Hyalomma aegyptium Ticks, Algeria
Figure
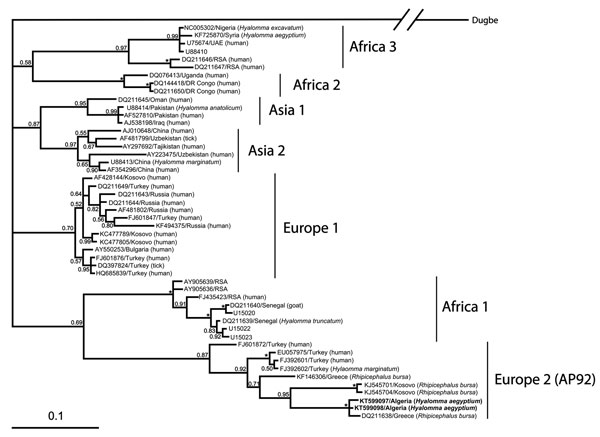
Figure. Phylogenetic analysis of Crimean-Congo hemorrhagic fever virus small RNA segment sequences, performed by using Bayesian inference in MrBayes version 3.1.2. (http://mrbayes.csit.fsu.edu/) under a general time-reversible plus gamma distribution plus invariable site model with 107 generations setup. Bootstrap values (>50%) are shown at nodes. Asterick (*) indicates 1.00 bootstrap value. Scale bar represents the estimated number of substitutions per site. Individual sequences are named with GenBank accession number/country of origin and the host, if available, in parentheses. Boldface indicates sequences of virus isolated from ticks collected from 12 Testudo graeca tortoises in Algeria, 2009–2010.
Page created: January 19, 2016
Page updated: January 19, 2016
Page reviewed: January 19, 2016
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.