Volume 22, Number 4—April 2016
Dispatch
Deletion Variants of Middle East Respiratory Syndrome Coronavirus from Humans, Jordan, 2015
Figure 2
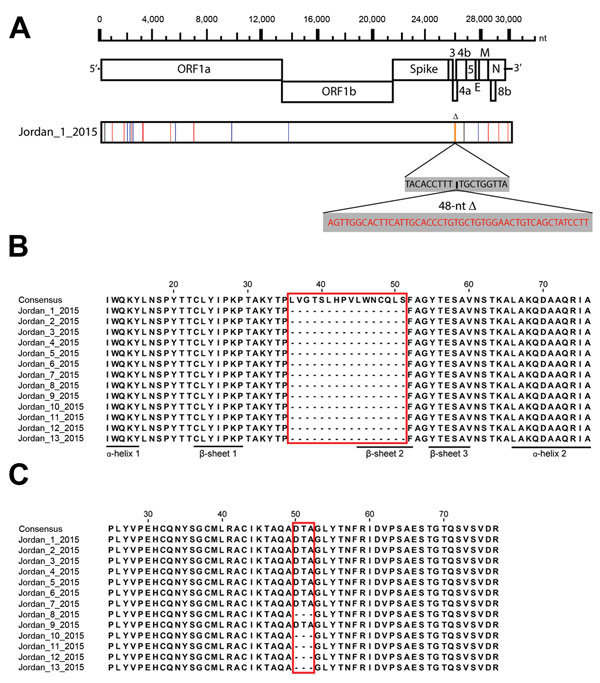
Figure 2. Genomic characterization of Middle East respiratory syndrome coronaviruses (MERS-CoVs) from Jordan. A) Nucleotide differences between MERS-CoV strain Jordan-1-2015 and the consensus sequence of the Riyadh 2015 cluster. Nucleotide changes are indicated in green (A), red (T), blue (C), and black (G). Deletions are indicated in orange. Deleted nucleotides are indicated in red. B) Protein sequence alignment of open reading frame (ORF) 4a (residues 10–76) of all 13 MERS-CoV strains from Jordan in 2015 compared with the consensus sequence of the Riyadh 2015 cluster. No nucleotide substitutions were observed in this region between MERS-CoVs from Jordan. Predicted secondary structures are indicated (13). Classical double-stranded RNA binding proteins have αβββα architecture. C) Protein sequence alignment of ORF3 (residues 25–77) of all 13 MERS-CoV strains from Jordan in 2015 compared with the consensus sequence of the Riyadh 2015 cluster. No nucleotide substitutions were observed in this region between MERS-CoVs from Jordan. Alignments were generated with the ClustalW program (http://pbil.univ-lyon1.fr/software/seaview) and visualized by using Jalview 2.9 (http://www.jalview.org/). Boxes in panels B and C indicate regions where amino acids have been deleted in the viruses from Jordan.
References
- Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–20. DOIPubMedGoogle Scholar
- World Health Organization. Emergencies preparedness, response: Middle East respiratory syndrome coronavirus (MERS-CoV) maps and epicurves [cited 2015 Nov 18]. http://www.who.int/csr/disease/coronavirus_infections/maps-epicurves/
- Drosten C, Meyer B, Müller MA, Corman VM, Al-Masri M, Hossain R, Transmission of MERS-coronavirus in household contacts. N Engl J Med. 2014;371:828–35. DOIPubMedGoogle Scholar
- Assiri A, McGeer A, Perl TM, Price CS, Al Rabeeah AA, Cummings DA, Hospital outbreak of Middle East respiratory syndrome coronavirus. N Engl J Med. 2013;369:407–16. DOIPubMedGoogle Scholar
- Cotten M, Watson SJ, Kellam P, Al-Rabeeah AA, Makhdoom HQ, Assiri A, Transmission and evolution of the Middle East respiratory syndrome coronavirus in Saudi Arabia: a descriptive genomic study. Lancet. 2013;382:1993–2002. DOIPubMedGoogle Scholar
- Drosten C, Muth D, Corman VM, Hussain R, Al Masri M. HajOmar W, et al. An observational, laboratory-based study of outbreaks of Middle East respiratory syndrome coronavirus in Jeddah and Riyadh, Kingdom of Saudi Arabia, 2014. Clin Infect Dis. 2015;60:369–77.
- Fagbo SF, Skakni L, Chu DKW, Garbati MA, Joseph M, Peiris M, Molecular epidemiology of hospital outbreak of Middle East respiratory syndrome, Riyadh, Saudi Arabia, 2014. Emerg Infect Dis [cited 2015 Oct 27]. DOIGoogle Scholar
- Seong MW, Kim SY, Corman VM, Kim TS, Cho SI, Kim MJ, Microevolution of outbreak-associated Middle East respiratory syndrome coronavirus, South Korea, 2015. Emerg Infect Dis [cited 2015 Nov 29]. DOIGoogle Scholar
- Chinese SARS Molecular Consortium. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303:1666–9. DOIPubMedGoogle Scholar
- World Health Organization. Global alert and response (GAR). Middle East respiratory syndrome coronavirus (MERS-CoV)—Jordan [cited 2015 Nov 18]. http://www.who.int/csr/don/01-september-2015-mers-jordan/en/
- Haagmans BL, Al Dhahiry SH, Reusken CB, Raj VS, Galiano M, Myers R, Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect Dis. 2014;14:140–5 and. DOIPubMedGoogle Scholar
- van Boheemen S, de Graaf M, Lauber C, Bestebroer TM, Raj VS, Zaki AM, Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio. 2012;3:e00473–12. DOIPubMedGoogle Scholar
- Siu KL, Yeung ML, Kok KH, Yuen KS, Kew C, Lui PY, Middle East respiratory syndrome coronavirus 4a protein is a double-stranded RNA-binding protein that suppresses PACT-induced activation of RIG-I and MDA5 in the innate antiviral response. J Virol. 2014;88:4866–76. DOIPubMedGoogle Scholar
- Niemeyer D, Zillinger T, Muth D, Zielecki F, Horvath G, Suliman T, Middle East respiratory syndrome coronavirus accessory protein 4a is a type I interferon antagonist. J Virol. 2013;87:12489–95. DOIPubMedGoogle Scholar
1These authors contributed equally to this article.