Volume 23, Number 1—January 2017
Dispatch
Norovirus Infection in Harbor Porpoises
Abstract
A norovirus was detected in harbor porpoises, a previously unknown host for norovirus. This norovirus had low similarity to any known norovirus. Viral RNA was detected primarily in intestinal tissue, and specific serum antibodies were detected in 8 (24%) of 34 harbor porpoises from the North Sea.
Noroviruses have been detected in humans, cats, dogs, pigs, sheep, cattle, sea lions, rodents, and bats (1–4). In humans, norovirus is a leading cause of gastroenteritis (1). Seven different norovirus genogroups have been described for the norovirus genus (1) that can be further subdivided in ≈30 genotypes. Noroviruses comprise a single-strand positive-sense RNA genome that is divided into 3 open reading frames (ORFs). Recombination among different genotypes is frequently observed for human noroviruses, most commonly near the junction of ORF1 and ORF2, leading to a recommendation for multilocus genotyping (5). Surprisingly, recombinant human noroviruses regularly contain previously undetected ORF1 sequences, which raises questions about the reservoirs of these viruses.
Noroviruses can spread through the fecal–oral route, and sewage contamination in coastal environments can result in contamination of shellfish, such as oysters. Oysters filter several liters of seawater daily and contain histo-blood group antigens resembling those of humans. These antigens can be specifically bound by noroviruses, resulting in bioaccumulation (6); as a result, eating oysters is linked to foodborne norovirus outbreaks in humans (7). This mode of transmission, however, could expose humans to viruses from other animal reservoirs, such as marine mammals, and vice versa.
A juvenile male harbor porpoise (Phocoena phocoena) ≈10.5 months of age was found alive on the coast of the Netherlands on May 10, 2012, and was transported to the SOS Dolphin Foundation (Harderwijk, the Netherlands) for rehabilitation (Technical Appendix). Important clinical signs at the rehabilitation center were anorexia, labored breathing, and disorientation. The animal showed no evidence of gastrointestinal disease, such as vomiting or diarrhea. Eight days after arrival in the rehabilitation center, the animal was euthanized because of the severity of clinical signs, and necropsy was performed according to standard procedures (8). The main pathology findings were bronchopneumonia associated with lungworm infection and encephalitis and hepatitis of unknown cause. The intestine did not show significant lesions macroscopically; microscopically, the enterocytes at the luminal surface of the intestine had sloughed into the lumen as a result of freeze–thaw artifact. The cells lining the intestinal crypts consisted of a mixture of enterocytes and mucus cells. The proportion of mucus cells increased progressively toward the end of the intestine. The lamina propria was infiltrated diffusely with a moderate number of lymphocytes, plasma cells, and eosinophils. This infiltrate was considered normal for this species.
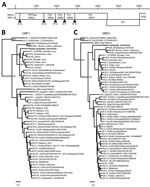
In the frame of a research program focusing on the identification of new viruses in possible reservoir hosts, we collected fecal material and performed random PCR in combination with 454-sequencing as described previously (9). This analysis resulted in 5,774 reads, of which 88 reads were most closely related to the norovirus genus, as determined by blastn and blastx analysis (10). Other reads that were most similar to viral genomes were most closely related to a coronavirus (16 reads [35%–94% nt identity]), salmon calicivirus (3 reads [11%–69% nt identity]), and porcine anello virus (1 read [91% nt identity], 2 reads [36% amino acid identity]). The harbor porpoise norovirus (HPNV) sequence was confirmed by Sanger sequencing (6,293 nt; GenBank accession no. KP987888) by using specific primers and comprising 3 ORFs, the partial ORF1 encoding the putative polyprotein, ORF2 encoding viral protein (VP) 1, and a partial ORF3 encoding VP2 (Figure 1, panel A). Phylogenetic analysis revealed that the HPNV RNA-dependent RNA-polymerase encoded by ORF1 clustered together with human genogroup I(GI) sequences (Figure 1, panel B), whereas VP1 clustered near strains belonging to human GI and bovine GIII (Figure 1, panel C).
To determine the tissue tropism of HPNV, we extracted RNA from formalin-fixed paraffin-embedded (FFPE) tissues collected from the HPNV-positive animal for histopathology. Tissues from all main organs and lymph nodes were tested for HPNV RNA by real-time PCR (Technical Appendix). Only the intestinal tissue and the FFPE material for immunohistochemical analysis (containing a mixture of tissues) were positive for norovirus with cycle threshold values of 30.5 and 37.4, respectively.
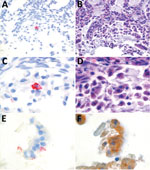
We conducted in situ hybridization to determine the cellular tropism of HPNV, as described previously (Figure 2) (12). HPNV-specific transcripts were detected in the cells in the intestine, indicating that this virus replicates in the intestinal tract. Sequential slides stained with hematoxylin and eosin or pankeratin showed that positive cells corresponded with enterocytes that had sloughed into the intestinal lumen because of freeze–thaw artifact, although we cannot exclude the possibility that other cell types were present (Figure 2).
To estimate the percentage of norovirus-infected harbor porpoises in our dataset, we extracted RNA from FFPE porpoise intestinal tissues collected from 48 animals during a 10-year period. Including the animal in which we detected HPNV, 10% (5/49) of the animals were positive for norovirus (cycle threshold <37) (Table 1), with primers designed to detect HPNV. Macroscopic and microscopic examination of the intestine of these animals showed no pathologic differences from harbor porpoises without norovirus infection.
To detect norovirus-specific antibodies in porpoise serum, we developed an ELISA based on HPNV VP1 and subsequently screened 34 harbor porpoise serum samples collected during 2006–2015 in the Netherlands (Technical Appendix). Samples from 8 (24%) harbor porpoises were positive for norovirus antibodies (Table 2). This dataset included samples from 2 harbor porpoises that were positive for norovirus RNA; however, their serum samples were negative for norovirus-specific antibodies.
Similar to human noroviruses, HPNV replicates in the intestine. B cells and enterocytes support human norovirus replication in vitro (13,14). We detected HPNV in cells corresponding to enterocytes, and it will be interesting to determine whether these viruses share receptor use with other noroviruses. In humans, norovirus infections are self-limiting in healthy persons but can result in illness and death in high-risk groups (4). Because the harbor porpoise in which we detected HPNV did not exhibit clinical signs of gastrointestinal disease, norovirus infection probably was not a major factor in the death of this animal.
Remarkably, the HPNV displayed 99% sequence homology to a short (300-nt) norovirus VP1 sequence detected in oysters (15). These oysters had been sampled because they were associated with a foodborne gastroenteritis outbreak in Ireland in 2012 (15). Oyster samples were collected from the restaurant where the outbreak occurred and from their harvesting area. Strains belonging to genotypes GI.1, GI.4, GII.4, GII.3, GII.1, GII.6, GI.2, GII.7, GI.11, and the strain that was homologous to HPNV were detected, although the HPNV-like strain was detected only in oysters from the harvesting area. In the patients, only GI.4, GI.2, GI.6, GII.1, and GII.7 strains were detected, but the fact that noroviruses infecting marine mammals closely related to human noroviruses have been found infecting harbor porpoises and contaminating oysters raises the question of whether HPNV could infect humans through contamination of oysters or other shellfish.
On the basis of our findings that norovirus infections might be a common infection in harbor porpoises from the southern North Sea and the detection of a norovirus in a sea lion (3), it is not unlikely that noroviruses are common in other marine mammals as well. The high genetic diversity within this genus complicates detection of new noroviruses. The discovery of HPNV and the recent discovery of noroviruses in bats highlight that much still remains to be discovered about animal reservoirs of noroviruses and triggers questions about the zoonotic potential of these viruses.
Dr. de Graaf is a research scientist at the Department of Viroscience, Erasmus Medical Center, in Rotterdam. Her primary research interests focus on noroviruses and evolution and determinants of host range.
Acknowledgments
We thank Peter van Run, Suzanne van Veen, and Lonneke Leijten for excellent technical assistance and SOS Dolphin Foundation for its assistance and support.
This work was supported by the European Union H2020 grant COMPARE under grant agreement no. 643476; the Virgo Consortium, funded by Dutch government project no. FES0908; and the ZonMw TOP grant under grant no. 91213058.
References
- Vinjé J. Advances in laboratory methods for detection and typing of norovirus. J Clin Microbiol. 2015;53:373–81. DOIPubMedGoogle Scholar
- Wu Z, Yang L, Ren X, He G, Zhang J, Yang J, et al. Deciphering the bat virome catalog to better understand the ecological diversity of bat viruses and the bat origin of emerging infectious diseases. ISME J. 2016;10:609–20. DOIPubMedGoogle Scholar
- Li L, Shan T, Wang C, Côté C, Kolman J, Onions D, et al. The fecal viral flora of California sea lions. J Virol. 2011;85:9909–17. DOIPubMedGoogle Scholar
- de Graaf M, van Beek J, Koopmans MP. Human norovirus transmission and evolution in a changing world. Nat Rev Microbiol. 2016;14:421–33. DOIPubMedGoogle Scholar
- Bull RA, Hansman GS, Clancy LE, Tanaka MM, Rawlinson WD, White PA. Norovirus recombination in ORF1/ORF2 overlap. Emerg Infect Dis. 2005;11:1079–85. DOIPubMedGoogle Scholar
- Maalouf H, Schaeffer J, Parnaudeau S, Le Pendu J, Atmar RL, Crawford SE, et al. Strain-dependent norovirus bioaccumulation in oysters. Appl Environ Microbiol. 2011;77:3189–96. DOIPubMedGoogle Scholar
- European Food Safety Authority. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2012. EFSA J. 2014;12:3547. DOIGoogle Scholar
- Kuiken T, Baker JR. Guidelines for the post mortem and tissue sampling of cetaceans. In: Law RJ, editor. Fisheries research technical report 97. Lowestoft (UK): Ministry of Agriculture, Fisheries and Food; 1994. p. 5–10.
- Bodewes R, Rubio García A, Wiersma LC, Getu S, Beukers M, Schapendonk CM, et al. Novel B19-like parvovirus in the brain of a harbor seal. PLoS One. 2013;8:e79259. DOIPubMedGoogle Scholar
- Schürch AC, Schipper D, Bijl MA, Dau J, Beckmen KB, Schapendonk CM, et al. Metagenomic survey for viruses in Western Arctic caribou, Alaska, through iterative assembly of taxonomic units. PLoS One. 2014;9:e105227. DOIPubMedGoogle Scholar
- Herfst S, van den Brand JMA, Schrauwen EJA, de Wit E, Munster VJ, van Amerongen G, et al. Pandemic 2009 H1N1 influenza virus causes diffuse alveolar damage in cynomolgus macaques. Vet Pathol. 2010;47:1040–7. DOIPubMedGoogle Scholar
- van Elk C, van de Bildt M, van Run P, de Jong A, Getu S, Verjans G, et al. Central nervous system disease and genital disease in harbor porpoises (Phocoena phocoena) are associated with different herpesviruses. Vet Res (Faisalabad). 2016;47:28. DOIPubMedGoogle Scholar
- Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, et al. Replication of human noroviruses in stem cell-derived human enteroids. Science. 2016;353:1387–93. DOIPubMedGoogle Scholar
- Jones MK, Watanabe M, Zhu S, Graves CL, Keyes LR, Grau KR, et al. Enteric bacteria promote human and mouse norovirus infection of B cells. Science. 2014;346:755–9. DOIPubMedGoogle Scholar
- Rajko-Nenow P, Keaveney S, Flannery J, McINTYRE A, Doré W. Norovirus genotypes implicated in two oyster-related illness outbreaks in Ireland. Epidemiol Infect. 2014;142:2096–104. DOIPubMedGoogle Scholar
Figures
Tables
Cite This Article1Current affiliation: Utrecht University, Utrecht, the Netherlands.
Table of Contents – Volume 23, Number 1—January 2017
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Miranda de Graaf, Erasmus Medical Center—Viroscience, Wytemaweg 80 Rotterdam 3015CN, the Netherlands
Top