Volume 23, Number 1—January 2017
Dispatch
Norovirus Infection in Harbor Porpoises
Figure 1
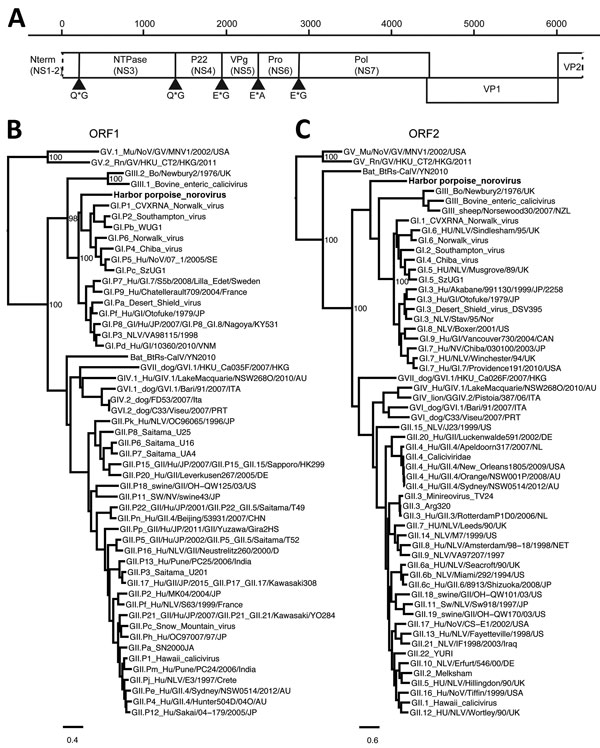
Figure 1. Genetic characterization of harbor porpoise norovirus. A) Genome organization of harbor porpoise norovirus. The putative cleavage sites are shown with arrowheads. B, C) Maximum-likelihood trees of the RNA-dependent RNA-polymerase (B) and ORF 2 (C) were inferred by PhyML 3.0 software (http://www.atgc-montpellier.fr/phyml/) by using the general time reversible nucleotide substitution model. Selected bootstrap values >70 are depicted. Scale bars indicate nucleotide substitutions per site. NS, nonstructural; NTPase, nucleoside triphosphatase; ORF, open reading frame; P, protein; Pol, polymerase; Pro, protease; VP, viral protein.
1Current affiliation: Utrecht University, Utrecht, the Netherlands.
Page created: December 14, 2016
Page updated: December 14, 2016
Page reviewed: December 14, 2016
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.