Volume 23, Number 10—October 2017
Research Letter
Spread of Chikungunya Virus East/Central/South African Genotype in Northeast Brazil
Abstract
We investigated an outbreak of exanthematous illness in Maceió by using molecular surveillance; 76% of samples tested positive for chikungunya virus. Genetic analysis of 23 newly generated genomes identified the East/Central/South African genotype, suggesting that this lineage has persisted since mid-2014 in Brazil and may spread in the Americas and beyond.
Dengue virus (DENV), Zika virus (ZIKV), and chikungunya virus (CHIKV) co-circulate in Brazil, are predominantly transmitted by Aedes aegypti mosquitoes, and cause similar clinical symptoms upon infection, complicating epidemiologic surveillance. Brazil harbors the highest diversity of CHIKV in the Americas; both the Asian and the East/Central/South African (ECSA) lineages circulate in the country (1). Despite high prevalence of CHIKV in Brazil (352,773 notified cases during January 2016–May 2017) and its widespread distribution (2), little is known about its transmission. We report a molecular and genomic investigation of an outbreak of CHIKV infection in Maceió, Alagoas state, Northeast Brazil.
During March 30–May 3, 2016, ≈12,000 patients visited 2 private hospitals in Maceió; roughly 70% of them had exanthematous illness symptoms compatible with DENV, CHIKV, or Zika virus infection. We analyzed 273 randomly chosen samples by using molecular diagnostics and virus discovery methods. The study was approved by the Faculty of Medicine from the University of São Paulo Review Board, and we obtained informed consent from all participants.
Analyzed samples were from patients who were on average 37 years of age (range 1–86 years); 175 (64%) were female, and 198 (73%) resided in Maceió municipality. Diagnostic tests for DENV, ZIKV, and CHIKV confirmed that 208 (76%) were positive for CHIKV RNA (Technical Appendix). In addition, 66 (24%) were positive for Zika virus RNA and 36 (13.2%) were co-infected with CHIKV and Zika virus, consistent with Zika virus circulation in Northeast Brazil in mid-2016 (3). We detected no DENV infections. Cycle threshold (Ct) values for CHIKV RNA–positive samples were lower (average Ct = 24.6) than those for ZIKV (average Ct = 33.5).
We applied a metagenomics next-generation sequencing protocol to 38 randomly chosen CHIKV RNA–positive samples (4) (Technical Appendix). We recovered 23 CHIKV genomes (>4,000 bp) by using the MiSeq Sequencer (Illumina, Inc., San Diego, CA, USA); mean genome coverage was 72× and mean depth coverage 207× (Technical Appendix Table 1). We also included 2 CHIKV RNA–positive samples from João Pessoa, Paraíba state. We did not detect the E1-A226V adaptive mutation associated with large outbreaks in Asia (5) in the strains we analyzed. We then appended sequences to publicly available data (659 CHIKV isolates) and used maximum-likelihood and Bayesian phylogenetic analysis to identify the origins of the outbreak (Technical Appendix).
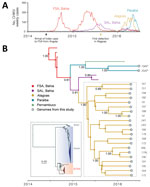
On the basis of available sequences of isolates from the Americas, the Maceió sequences we analyzed fell within a single strongly supported monophyletic clade (bootstrap support = 99%, posterior support = 1.00) that belongs to the ECSA genotype (Figure). Genetic analysis suggests the outbreak most likely originated from transmission cycles not previously identified in Northeast Brazil and not from a separate introduction into the Americas. Before August 2015, no CHIKV infections had been reported in Alagoas (Figure). Molecular dating analysis indicates that the outbreak was caused by a single founder strain that is estimated to have arrived in Alagoas around late April 2015 (95% Bayesian credible interval July 2014–October 2015), possibly a few months before the earliest reports of CHIKV there (Figure). Our reconstruction of the history of the ECSA genotype in Brazil using a phylogeographic approach (6) further suggests that this lineage was introduced into Alagoas from the neighboring Bahia state, which experienced a CHIKV epidemic during January–August 2015 (7).
The brief to negligible period of undetected transmission of CHIKV in Alagoas is consistent with past reports (8) and in contrast with the unrecognized circulation in the region of Zika virus, which typically causes milder symptoms (3). The most common clinical signs and symptoms for the sequenced CHIKV cases were fever (87%), arthralgia (70%), headache (44%), exanthema (30%), and myalgia (26%) (Technical Appendix Table 2). CHIKV infection is often characterized by prolonged periods of disability. Further investigation is needed to study potential differences in the effects of CHIKV and Zika virus infection on public health, as well in pathology and innate and adaptive immune responses to each genotype.
The unrecognized transmission of the CHIKV ECSA genotype in Northeast Brazil is unique in the Americas. The spread of this genotype in this region will be mediated by several factors, including herd immunity, vector suitability, and human mobility. Serologic and molecular surveys in human and mosquito populations are required to characterize the factors involved in transmission and the extent of cross-protection of the Asian and the ECSA genotypes in the Americas. Although CHIKV ECSA has been found only in Ae. aegypti mosquitoes (9), a recent study has shown that Ae. albopictus mosquitoes in Brazil are also highly competent in CHIKV ECSA transmission (10). Given the widespread distribution of both vectors in the Americas (1), it is possible that the ECSA lineage may spread to other regions in the Americas and beyond. A better understanding of the transmission dynamics of CHIKV, DENV, and Zika virus in the Americas is essential to fully understand the risk of arbovirus-associated congenital anomalies, Guillain-Barré, and other neurological syndromes.
Dr. da Costa is a postdoctoral researcher at the Institute of Tropical Medicine, University of São Paulo, Brazil. He is interested in pathogen discovery. Dr. Thézé is a postdoctoral researcher at the University of Oxford, UK. He is interested in viral evolution, especially the spatiotemporal dynamics of pathogen spread.
Acknowledgments
We thank all participants in this study and Luciano Monteiro da Silva Patricia Chefre, Renata Palma, and Suzana Santos for support. We also thank Illumina, Inc., Sage Science, Inc., Promega Biotecnologia do Brasil, Ltda, and Greiner Bio-One Brasil Produtos Médicos Hospitalares Ltda for the donation of reagents and plastics for this project.
This work was supported by FAPESP #2016/01735-2 and CNPq #400354/2016-0. A.C.dC. is funded by FAPESP #2012/03417-7 and 2017/00021-9. N.R.F. is funded by a Sir Henry Dale Fellowship (Wellcome Trust/Royal Society Grant 204311/Z/16/Z). This research received funding from the ERC under the European Union’s Seventh Framework Programme (FP7/2007-2013), grant agreement no. 614725-PATHPHYLODYN; and the US Agency for International Development Emerging Pandemic Threats Program-2 PREDICT-2 (Cooperative Agreement No. AID-OAA-A-14-00102).
References
- Nunes MR, Faria NR, de Vasconcelos JM, Golding N, Kraemer MU, de Oliveira LF, et al. Emergence and potential for spread of Chikungunya virus in Brazil. BMC Med. 2015;13:102. DOIPubMedGoogle Scholar
- Ministério da Saúde. Boletins Epidemiológicos—Secretaria de Vigilância em Saúde. Situação epidemiológica / dados dengue. 2017;48:16 [cited 2017 Jul 13] http://portalsaude.saude.gov.br/index.php/situacao-epidemiologica-dados-dengue
- Faria NR, Quick J, Claro IM, Thézé J, de Jesus JG, Giovanetti M, et al. Establishment and cryptic transmission of Zika virus in Brazil and the Americas. Nature. 2017;546:406–10. DOIPubMedGoogle Scholar
- Deng X, Naccache SN, Ng T, Federman S, Li L, Chiu CY, et al. An ensemble strategy that significantly improves de novo assembly of microbial genomes from metagenomic next-generation sequencing data. Nucleic Acids Res. 2015;43:e46. DOIPubMedGoogle Scholar
- Tsetsarkin KA, Vanlandingham DL, McGee CE, Higgs S. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog. 2007;3:e201. DOIPubMedGoogle Scholar
- Lemey P, Rambaut A, Drummond AJ, Suchard MA. Bayesian phylogeography finds its roots. PLOS Comput Biol. 2009;5:e1000520. DOIPubMedGoogle Scholar
- Cardoso CW, Kikuti M, Prates AP, Paploski IAG, Tauro LB, Silva MMO, et al. Unrecognized emergence of chikungunya virus during a Zika virus outbreak in Salvador, Brazil. PLoS Negl Trop Dis. 2017;11:e0005334. DOIPubMedGoogle Scholar
- Faria NR, Lourenco J, Cerqueira EM, Lima MM, Pybus OG, Alcantara LCJ. Epidemiology of chikungunya virus in Bahia, Brazil, 2014–2015. PLoS Currents Outbreaks. 2016;1 [cited 2007 Jul 13] http://currents.plos.org/outbreaks/article/epidemiology-of-chikungunya-virus-in-bahia-brazil-2014-2015
- Costa-da-Silva AL, Ioshino RS, Petersen V, Lima AF, Cunha MDP, Wiley MR, et al. First report of naturally infected Aedes aegypti with chikungunya virus genotype ECSA in the Americas. PLoS Negl Trop Dis. 2017;11:e0005630. DOIPubMedGoogle Scholar
- Vega-Rúa A, Zouache K, Girod R, Failloux A-B, Lourenço-de-Oliveira R. High level of vector competence of Aedes aegypti and Aedes albopictus from ten American countries as a crucial factor in the spread of Chikungunya virus. J Virol. 2014;88:6294–306. DOIPubMedGoogle Scholar
Figure
Cite This Article1These authors contributed equally to this article.
Table of Contents – Volume 23, Number 10—October 2017
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Nuno R. Faria, Peter Medawar Building, South Parks Road, OX1 3SY Oxford, UK; ; Ester C. Sabino, Instituto de Medicina Tropical, Universidade de São Paulo, Avenida Dr. Enéas Carvalho de Aguiar, 470- CEP 05403-000, São Paulo, Brazil
Top