Volume 23, Number 11—November 2017
Dispatch
Phylogenetic Analysis of Klebsiella pneumoniae from Hospitalized Children, Pakistan
Cite This Article
Citation for Media
Abstract
Klebsiella pneumoniae shows increasing emergence of multidrug-resistant lineages, including strains resistant to all available antimicrobial drugs. We conducted whole-genome sequencing of 178 highly drug-resistant isolates from a tertiary hospital in Lahore, Pakistan. Phylogenetic analyses to place these isolates into global context demonstrate the expansion of multiple independent lineages, including K. quasipneumoniae.
Klebsiella spp. are gram-negative bacteria that are widely distributed in the environment, and K. pneumoniae is a common cause of infection in humans (1). Increasingly, K. pneumoniae is reported as a cause of invasive bloodborne infections, particularly in healthcare settings and in immunocompromised patients (2). Of concern is that infection-associated K. pneumoniae is often multidrug resistant (MDR) and can harbor resistance determinants against most, if not all, commonly used antimicrobial drugs, posing a major threat to public health. The World Health Organization recently highlighted finding new treatments against MDR Enterobacteriaceae (including Klebsiella) as priority 1 (critical) (http://www.who.int/mediacentre/news/releases/2017/bacteria-antibiotics-needed/en/).
K. pneumoniae is a major pathogen in economically developed settings, and multiple outbreaks in different countries have been reported. Less is known about its prevalence in economically challenged areas, including lower and middle income countries (LMIC). Reports are now appearing about Klebsiella-associated infections in Nepal (3) and in Indonesia, Laos, and Vietnam (1). Klebsiella can spread rapidly in hospital environments, and the increasing prevalence of MDR strains has raised concern among major health organizations (4,5). Thus, high-resolution insight into the diversity of Klebsiella spp. isolated in LMICs will provide vital data for improving epidemiologic management of infections and for better understanding of the mechanisms of spread between LMICs and more developed countries.
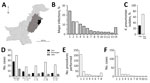
Clinical samples were collected during a 22-month period (May 2010–February 2012) from The Children’s Hospital & The Institute of Child Health, Lahore (Lahore, Pakistan), the largest tertiary care hospital in the region (Figure 1, panel A). The hospital had a capacity of 650 beds during the study period but is under pressure to handle up to 2,000 inpatients at any given time. The primary catchment area is Lahore (population ≈10 million); the hospital also receives patients from the greater area of Punjab province (population ≈100 million) (Figure 1, panel A). The Ethical Committee of The Children’s Hospital & Institute of Child Health, Lahore, approved the study.
A total of 44,260 samples were collected in the course of routine sampling from children; 5,475 (12.4%) resulted in laboratory-positive cultures. Of these, 710 (13.0%) samples were positively identified as K. pneumoniae, the third most dominant isolate after Escherichia coli (1,336 [24.4%]) and coagulase-negative staphylococci (724 [13.2%]) (Figure 1, panel B). We screened all K. pneumoniae isolates for resistance to ceftazidime (30 μg disc, zone of inhibition <17 mm) or cefotaxime (30 μg disc, zone of inhibition <22 mm). We further tested K. pneumoniae isolates that were resistant to any of these indicator drugs using the Clinical and Laboratory Standards Institute combined-disc confirmatory test (6); extended-spectrum β-lactamase (ESBL) production was confirmed when the zone of inhibition by either cephalosporin drug increased by >5 mm in the presence of clavulanate. A total of 214 of K. pneumoniae isolates were ESBL-positive (Figure 1, panel C); most were isolated from children with bloodstream infections (Figure 1, panel D). The outcomes were severe, especially among neonatal patients (Figure 1, panel D); 56 died, 31 were taken home against medical advice, and 127 were discharged (Figure 1, panel D). Almost all patients infected with ESBL Klebsiella had received an intravenous line (209 [97.7%]) (Figure 1, panel E,F), and a high number received a urinary catheter (46 [21.5%]).
We performed whole-genome sequencing on 178 isolates (Technical Appendix Table 1). We prepared Illumina sequencing libraries (Illumina, San Diego, CA, USA) with a 450-bp insert size according to the manufacturer’s protocols and sequenced them on an Illumina HiSeq2000 with 100-bp–long paired-end reads before assembly using an open-source high-throughput assembly and improvement pipeline as described (7) (https://github.com/sanger-pathogens/) and annotated using prokka (8). Initial clustering using mash (9) enabled aligning of these isolates to published reference sequences (Technical Appendix Figure 1, panel A). The clustering indicated a strong structure for the isolates that fell within the species K. pneumoniae (Technical Appendix Figure 1, panel B). However, the analysis also revealed a large group of sequences most similar to K. quasipneumoniae; closer inspection focusing on this species showed strongest similarity to subspecies similipneumoniae (Technical Appendix Figure 1, panel C) (10). We combined several independent datasets: a large global collection (1); 2 hospital outbreaks obtained in a comparable time frame, 1 of which was based in Nepal in 2012 (3); and a hospital study from Spain that also focused on diversity within ESBL-producing strains (11) (Technical Appendix Table 2). We applied the pan-genome pipeline Roary version 3.7.0 (12) with a blastp (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins) percentage identity of 90% and a core definition of 99%, resulting in a core gene alignment comprising 1,793 genes for all studies (Figure 2) and 3,486 genes for the strains of this study (Technical Appendix Figure 2). We first extracted single-nucleotide polymorphisms using snp-sites version 2.3.2 (13), then calculated a maximum-likelihood tree using RAxML version 8.2.8 (14) with the general time-reversible model and 100 bootstrap repeats. The core gene phylogeny (Figure 2) shows a wide distribution of the isolates from Pakistan across different lineages rather than 1 clonal lineage. The diversity of our strain collection is further emphasized through the diversity of multilocus sequence types (STs). No single ST dominates (Figure 2 outer ring; Technical Appendix Figure 2); however, a large group of isolates belongs to ST15, which is known to be problematic. The presence of K. quasipneumoniae isolates agrees with an overall lower percentage of reads mapped against K. pneumoniae (Technical Appendix Table 1) and with recent descriptions of virulent K. quasipneumoniae strains (1,9,15). Assessing the metadata in phylogenetic context highlights the association of the K. quasipneumoniae lineage with patients in the neonatal ward, suggestive of its nosocomial residency (Technical Appendix Figure 2). However, other main lineages (e.g., ST15, ST48) show a dynamic spread across wards and age groups, indicating against >1 resident lineages but instead a frequent movement of K. pneumoniae through the hospital, general population, or both.
The high number of K. quasipneumoniae isolates, even if potentially restricted to most sequences derived from a lineage potentially resident in a specific ward, highlights the importance of a diverse set of sampling sites to be studied. It also highlights the need for continued monitoring of new emerging strains and that our knowledge of the diversity of potentially problematic lineages is far from exhaustive.
The Klebsiella isolates in this study represented the Klebsiella isolates routinely present in infections over a protracted period. Our findings highlight a consistent problem with ESBL-encoding strains belonging to a multitude of lineages. We observed sporadic single-isolate lineages, as well as smaller, related clusters of 5–10 strains per lineage, in addition to 2 larger clusters of strains. More studies are needed to better delineate the distinguishing features for successful spread and persistence of lineages such as the ST15 cluster. Also, the large spread of K. quasipneumoniae is unusual. Further intense monitoring of LMIC hospital environments is urgently needed to prevent the persistence of resident lineages with very high base-level drug resistance, which, through the inevitable acquisition of a few more genes, would lead to untreatable infections.
Dr. Ejaz is a microbiologist who is working as Assistant Professor in Aljouf University, Aljouf, Saudi Arabia. He also worked at The Children’s Hospital Lahore, Pakistan, and University of Melbourne, Australia. His primary research interests include medical bacteriology.
Acknowledgment
This work was supported by National Health and Medical Research Council program grants (0606788 to R.A.S. and T. L.; 1092262 to R.A.S., G.D., and T.L.); the Wellcome Trust (206194); and the Higher Education Commission of Pakistan and The Children’s Hospital & The Institute of Child Health, Lahore, Pakistan. H.E. was supported by a scholarship from Higher Education Commission Pakistan under the International Research Support Initiative Program.
References
- Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A. 2015;112:E3574–81. DOIPubMedGoogle Scholar
- Lee CR, Lee JH, Park KS, Kim YB, Jeong BC, Lee SH. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front Microbiol. 2016;7:895. DOIPubMedGoogle Scholar
- Chung The H, Karkey A, Pham Thanh D, Boinett CJ, Cain AK, Ellington M, et al. A high-resolution genomic analysis of multidrug-resistant hospital outbreaks of Klebsiella pneumoniae. EMBO Mol Med. 2015;7:227–39. DOIPubMedGoogle Scholar
- World Health Organization. Antimicrobial resistance: global report on surveillance 2014. Geneva: The Organization; 2014.
- Centers for Disease Control and Prevention. New carbapenem-resistant Enterobacteriaceae warrant additional action by healthcare providers [cited 2017 Jan 5]. https://emergency.cdc.gov/han/han00341.asp
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; twentieth informational supplement. M100-S20. Wayne (PA): The Institute; 2010.
- Page AJ, De Silva N, Hunt M, Quail MA, Parkhill J, Harris SR, et al. Robust high-throughput prokaryote de novo assembly and improvement pipeline for Illumina data. Microb Genom. 2016;2:e000083. DOIPubMedGoogle Scholar
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. DOIPubMedGoogle Scholar
- Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016;17:132. DOIPubMedGoogle Scholar
- Brisse S, Passet V, Grimont PA. Description of Klebsiella quasipneumoniae sp. nov., isolated from human infections, with two subspecies, Klebsiella quasipneumoniae subsp. quasipneumoniae subsp. nov. and Klebsiella quasipneumoniae subsp. similipneumoniae subsp. nov., and demonstration that Klebsiella singaporensis is a junior heterotypic synonym of Klebsiella variicola. Int J Syst Evol Microbiol. 2014;64:3146–52. DOIPubMedGoogle Scholar
- Pérez-Vázquez M, Oteo J, García-Cobos S, Aracil B, Harris SR, Ortega A, et al. Phylogeny, resistome and mobile genetic elements of emergent OXA-48 and OXA-245 Klebsiella pneumoniae clones circulating in Spain. J Antimicrob Chemother. 2016;71:887–96. DOIPubMedGoogle Scholar
- Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3. DOIPubMedGoogle Scholar
- Page AJ, Taylor B, Delaney AJ, Soares J, Seemann T, Keane JA, et al. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb Genom. 2016;2:e000056. DOIPubMedGoogle Scholar
- Stamatakis A. Using RAxML to infer phylogenies. Curr Protoc Bioinformatics. 2015;51:6.14.1-14.
- Arena F, Henrici De Angelis L, Pieralli F, Di Pilato V, Giani T, Torricelli F, et al. Draft genome sequence of the first hypermucoviscous Klebsiella quasipneumoniae subsp. quasipneumoniae isolate from a bloodstream infection. Genome Announc. 2015;3:e00952–15. DOIPubMedGoogle Scholar
Figures
Cite This Article1These authors contributed equally to this article.
2Current affiliation: CAMS, Aljouf University, Aljouf, Saudi Arabia.
Table of Contents – Volume 23, Number 11—November 2017
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Richard A. Strugnell, Department of Microbiology and Immunology, The University of Melbourne, at Peter Doherty Institute of Infection and Immunity, Melbourne, VIC, Australia; ; Eva Heinz, Infection Genomics Program, Wellcome Trust Sanger Institute, Hinxton, CB10 1SA, UK
Top