Volume 23, Number 12—December 2017
Research
Characterization of Streptococcus pyogenes from Animal Clinical Specimens, Spain
Cite This Article
Citation for Media
Abstract
Streptococcus pyogenes appears to be almost exclusively restricted to humans, with few reports on isolation from animals. We provide a detailed characterization (emm typing, pulsed-field gel electrophoresis [PFGE], and multilocus sequence typing [MLST]) of 15 S. pyogenes isolates from animals associated with different clinical backgrounds. We also investigated erythromycin resistance mechanisms and phenotypes and virulence genes. We observed 2 emm types: emm12 (11 isolates) and emm77 (4 isolates). Similarly, we observed 2 genetic linages, sequence type (ST) 26 and ST63. Most isolates exhibited the M macrolide resistance phenotype and the mefA/ermB genotype. Isolates were grouped into 2 clones on the basis of emm-MLST-PFGE-virulence gene profile combinations: clone 1, characterized by the combined genotype emm12-ST36-pulsotype A-speG; and clone 2, characterized by the genotype emm77-ST63-pulsotype B-speC. Our results do not show conclusively that animals may represent a new reservoir of S. pyogenes but indicate the ability of human-derived S. pyogenes isolates to colonize and infect animals.
Streptococcus pyogenes (group A Streptococcus) is a gram-positive bacterium that causes several diseases in humans. S. pyogenes usually colonizes the throat or skin epithelial surfaces and causes a wide variety of clinical manifestations, such as noninvasive pharyngitis, dermatitis, and scarlet fever (1,2). However, this pathogen is also responsible for deadly invasive systemic infections such as necrotizing fasciitis and streptococcal toxic shock syndrome (3).The ecologic niche of S. pyogenes appears to be quite narrow, with humans being the almost exclusive biologic host (4) and no animal or environmental reservoir of known importance contributing to its life cycle (2). Reports of isolation of S. pyogenes from sources other than humans are rare. S. pyogenes has recently been associated with an infection in a free-living European hedgehog (Erinaceus europaeus) (5). S. pyogenes has also been recovered from the feces of a dog with possible antibiotic-associated colitis (6) and from the eye discharge of a dog with conjunctivitis (7). We know of no other reports of isolation of this microorganism from animals.
We conducted a study to provide a detailed characterization of animal S. pyogenes isolates using emm typing, pulsed-field gel electrophoresis (PFGE), and multilocus sequence typing (MLST). We also investigated erythromycin resistance mechanisms and phenotypes, as well as virulence genes.
Origin and Identification of Bacterial Isolates
We analyzed 15 isolates of S. pyogenes obtained from rabbits (n = 14) and sheep (n = 1) in Spain during 2006–2014 (Table 1). Most rabbit isolates were from unrelated animals, located in different commercial farms (n = 14) and locations throughout Spain. Links between rabbit farms were not identified. The sheep included in this study was from a farm that had no rabbits. Human contact with animals was restricted to the personnel working in the rabbit farms and sheep flocks.
We recovered isolates from different clinical backgrounds: 8 from skin infections, 4 from genital tract infections, and 1 each from respiratory infections, mastitis, and otitis. We collected samples from skin and ear infections with sterile cotton swabs and collected the milk sample from the mastitis case aseptically in a sterile tube. Rabbits with genital tract or lung infections were euthanized, at farms or laboratories, and necropsied under aseptic conditions; clinical specimens were collected with forceps and scissors scrubbed in 70% ethanol. Samples taken at farms were transported to the laboratory in refrigerated polyethylene bags and processed within 24 hours after sampling.
Clinical specimens were sampled onto blood agar plates that were incubated at 37°C for 24–48 hours. Identification of isolates as S. pyogenes was based on colony morphology, β-hemolysis, and biochemical characteristics using the commercial identification system rapid ID 32 STREP (BioMerieux, Marcy L’Étoile, France). Biochemical identification was also confirmed by sequencing the 16S rRNA gene (8).
Antimicrobial Drug Susceptibility Tests
We performed drug susceptibility testing using the Clinical and Laboratory Standards Institute broth microdilution method (9) in Mueller–Hinton broth supplemented with 5% lysed horse blood. We determined the susceptibilities of the isolates with a commercially available susceptibility test (CMV3AGPF Sensititer standard panel; Trek Diagnostics, West Essex, UK) performed according to the manufacturer’s instructions. The agents we tested were penicillin (0.25–16 μg/mL), erythromycin (0.25–8 μg/mL), vancomycin (0.25–32 μg/mL), daptomycin (0.25–16 μg/mL), chloramphenicol (2–32 μg/mL), linezolid (0.5–8 μg/mL), tetracycline (1–32 μg/mL), quinupristin (0.5–32 μg/mL), tigecycline (0.05–0,5 μg/mL), streptomycin (512–2048 μg/mL), kanamycin (128–1024 μg/mL), lincomycin (1–8 μg/mL), and gentamicin (128–1024 μg/mL). In addition, we determined MICs of clindamycin, erythromycin, and tetracycline by Etest (AB Biodisk, Solna, Sweden). We interpreted the results using the Clinical and Laboratory Standards Institute breakpoints for streptococci (9) for penicillin, erythromycin, vancomycin, daptomycin, chloramphenicol, tetracycline, and quinupristin; the European Committee on Antimicrobial Susceptibility Testing breakpoints for tigecycline and linezolid (http://www.eucast.org/clinical_breakpoints); and the Comité de l’Antibiogramme de la Société Française de Microbiologie breakpoints (10) for streptomycin, kanamycin, lincomycin, and gentamicin.
Macrolide Resistance Phenotype
To identify macrolide resistance phenotypes, we used a double-disk diffusion test (D-zone test) using erythromycin (15 μg) and clindamycin (2 μg) disks, as described by Hasenbein et al. (11). Isolates with blunting of the clindamycin inhibition zone around the disk adjacent to the erythromycin disk were considered to have an iMLSB phenotype (erythromycin resistant and clindamycin inducible). Clindamycin-susceptible isolates without blunting indicated an M phenotype (erythromycin resistant and clindamycin susceptible). Isolates that were resistant to both antimicrobial drugs were considered to have a cMLSB phenotype (constitutive erythromycin and clindamycin resistant).
Detection of Macrolides and Tetracycline Resistance Genes
We extracted DNA according to the protocol in the US Centers for Disease Control and Prevention (CDC) S. pyogenes sequence database (http://www.cdc.gov/ncidod/biotech/strep/protocols.htm). We screened all erythromycin-resistant isolates by PCR for the erythromycin resistance genes ermB (12), ermA (13), mefA (14), and msrD (15). We tested tetracycline-resistant isolates for the tetracycline resistance genes tetM and tetO (14).
Detection of Virulence Genes
We tested the S. pyogenes isolates for the presence of the virulence genes speA, speB, speC, speF, speG, speH, speJ, speM, ssa, and smeZ by PCR. We used primers and conditions described previously (16,17).
PFGE Analysis, MLST, and emm Typing
For PFGE analysis, genomic DNAs of the S. pyogenes isolates were prepared and digested with SmaI restriction enzyme (MBI Fermentas, Vilnius, Lithuania) following a previously published protocol (18). We performed MLST following the method established by Enright et al. (19) and assigned the allele and sequence type (ST) according to the PubMLST website (http://pubmlst.org/spyogenes). We amplified and sequenced the emm gene according to the protocol of the CDC International Streptococcal Reference Laboratory (http://www.cdc.gov/streplab/protocol-emm-type.html). We compared the sequences of the emm genes with those in the CDC database using BLAST analysis (http://www.cdc.gov/ncidod/biotech/strep/strepblast.htm) for type assignment.
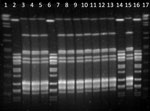
We observed 2 emm types (Table 2): emm12 was the most frequent (11 isolates), followed by emm77 (4 isolates). Two pulsotypes (A and B) were generated after typing the isolates by PFGE with the restriction enzyme SmaI; 11 isolates were pulsotype A and 4 isolates pulsotype B (Figure). Similarly, we observed 2 genetic linages (ST26 and ST63) after MLST analysis.
All 15 S. pyogenes isolates were susceptible to penicillin (MIC <0.25 mg/L), vancomycin (MICs <0.25 to 0.5 mg/L), daptomycin (MIC <0.25 mg/L), chloramphenicol (MICs <2 to 4 mg/L), tigecycline (MICs <0.015 to 0.12 mg/L), and gentamicin (MIC <128 mg/L). Additionally, all isolates but 1 were susceptible to kanamycin (MIC <128 mg/L), and 12 isolates showed susceptibility to linezolid (MICs <2 mg/L), streptomycin (MICs >2,048 mg/L), and lincomycin (>8 mg/L). On the other hand, all isolates were resistant to tetracycline, with MICs ranging from 24 to 96 mg/L using Etest (Table 2). Eleven isolates showed tetracycline-resistant genotype tetM/tetO, 2 isolates tetO, and 1 isolate tetM (Table 2).
Most isolates (7/15) exhibited the M phenotype, 2 isolates the phenotype cMLSB, and 1 the phenotype iMLSB (Table 2). The macrolide-resistant genotype mefA/ermB was the most frequently observed, seen in all isolates but 1 with the M phenotype and in the isolate with phenotype cMLSB. The genotype ermB was observed alone in 1 isolate of each phenotype. No isolate carried the msrD or ermA macrolide-resistant determinants.
We detected the chromosomal-encoded speB and speF genes in all isolates. We observed 2 different virulence gene profiles based on the presence/absence of the speG and speC genes. We detected the genotype speG in 11 isolates and the genotype speC in 4 isolates (Table 2).
We grouped the 15 S. pyogenes isolates into 2 different clones on the basis of emm-MLST-PFGE-virulence genes profile combinations. Clone 1 grouped isolates characterized by the combined genotype emm12-ST36-pulsotype A-speB/speF/speG, whereas isolates of clone 2 were characterized by the genotype emm77-ST63-pulsotype B-speB/speF/speC (Table 2). In addition, isolates of clone 1 were erythromycin resistant, mainly exhibiting an M phenotype, and isolates of clone 2 were erythromycin susceptible.
S. pyogenes is a human pathogen that has rarely been isolated from animals. It has been isolated from abscesses in cervical and mesenteric lymph nodes and liver of a free-living European hedgehog (E. europaeus) and from 2 dogs with severe colonic disease and conjunctivitis (5–7). Here we describe the detailed characterization of animal S. pyogenes isolates from different clinical specimens obtained from rabbits (n = 14) and sheep (n = 1) in Spain during 2006–2014. This pathogen was recovered mainly from noninvasive cases, with skin infections being the most common clinical presentation (n = 6), followed by genital tract infections (n = 4) (Table 1). S. pyogenes was isolated from all skin clinical samples together with Staphylococcus aureus, a well-recognized pathogen associated with different skin diseases in animals (20). These results indicate that although S. pyogenes should be able to colonize the skin of animals, it is difficult to ascertain its etiologic significance in skin infections. However, S. pyogenes was isolated in pure culture from clinical specimens of the genital tract, ears, mammary glands, and lungs in rabbits, indicating the potential role of S. pyogenes in these infections.
Most of the S. pyogenes isolates we tested (n = 11) exhibited the genotype emm12-ST36, which has been isolated repeatedly from humans in different countries (21–27), including Spain (28–30). This genotype can exhibit an M phenotype (31) and has been associated with skin and soft tissue infections (32), data that fit with our results, as more than half of the isolates with this genotype were isolated from abscesses and dermatitis (Table 2). The genotype emm77-ST63 that we identified in 4 animal isolates has also been detected in human S. pyogenes isolates (21,25,33), but unlike human isolates, the isolates in our study were erythromycin and clindamycin susceptible (Table 2).
All 11 isolates in clone 1 (pulsotype A) exhibited PFGE profiles that were indistinguishable from each other, and all 4 isolates in clone 2 also exhibited PFGE profiles that were indistinguishable PGFE from each other (pulsotype B; Figure). Isolates of S. pyogenes usually exhibit high levels of genetic diversity (4). Thus, the fact that we identified only 2 clones in different isolates collected over a period of 8 years was unexpected. The possibility of a common source of infection is very unlikely because all isolates were recovered at different times from different animals in farms located at geographically distant locations spread throughout Spain, without any epidemiologic relationship (Table 1). In addition, clinical specimens were processed independently in the same laboratory by highly qualified and trained personnel, which makes the possibility of a cross-contamination in the laboratory unlikely.
Under these conditions, multiple human-to-animal transmission events should be the most likely origin of these genotypes in sheep and rabbits. Another possible explanation could be that genotypes ST36 and ST63, although originating from humans, represent genetic linages with a specific host tropism, mainly for rabbits, which contributed to their successful dissemination in these animals, as observed with other streptococci (34). Cases of S. pyogenes infection were not recorded among the personnel working in the rabbit farms and sheep flock from which S. pyogenes was isolated. Asymptomatic human carriers have a key role in S. pyogenes transmission (35). For these reasons and even though screenings to identify asymptomatic S. pyogenes carriers were not carried out, we can speculate that asymptomatic employees were the most probable source of S. pyogenes in the animals included in the study. Although we cannot infer from the results of this study that animals, mainly rabbits, may represent a new reservoir of S. pyogenes, the results clearly indicate the ability of human-derived S. pyogenes isolates to colonize and infect animals, which could be more frequent than has been recognized until now.
Isolates with the genotype mefA/ermB usually correlate with the cMLSB phenotype, but 5 of the 6 S. pyogenes isolates with the mefA/ermB genotype in our study exhibited M phenotype (Table 2), which agrees with previous observations (29). The erm gene usually confers co-resistance to macrolides, lincosamides, and streptogramins. Curiously, all M phenotype isolates in our study showed susceptibility to clindamycin and were positive for the emrB gene. This result, although unusual, has also been observed previously in S. pyogenes isolates from different countries (26,36–38). A possible explanation could be that the ermB gene was nonfunctional in the isolates with clindamycin-susceptible phenotypes. The isolate M72193 exhibited the iMLSB phenotype but was ermA-negative (Table 2). This result, although infrequent, has also been observed in previous studies (39). Isolates with the iMLSB phenotype have been further subdivided into 3 distinct types: type A, associated with the presence of the ermB gene; and types B and C, associated with the presence of the ermA gene (40,41). This isolate carried the ermB gene (Table 2), suggesting therefore an iMLSB-A phenotype.
Unlike most human S. pyogenes isolates, which usually carry either tetM or tetO genes, most of the isolates in this study (n = 11) carried both genes (Table 2). Human isolates with the combination of tetM and tetO tetracycline-resistance genes have been identified previously in Spain (29). Another uncommon result was the identification of 1 isolate (83553) that was resistant to tetracycline (MIC 64 mg/L) but lacked resistance tetM and tetO genes (Table 2) commonly associated with tetracycline resistance in S. pyogenes (42). However, tetracycline-resistant strains and negativity to these genes have also been reported (43). Further studies will be necessary to elucidate the precise mechanism of resistance to tetracycline in this strain.
In summary, this study provides a detailed characterization of animal S. pyogenes isolates associated with different clinical backgrounds. This pathogen should be considered by veterinary microbiologists when processing clinical material from animals.
Dr. Vela is an associate professor at the Animal Health Department, Veterinary Faculty, Complutense University, Madrid, Spain. Her research focuses on the characterization of relevant animal bacterial pathogens.
References
- Bisno AL, Stevens D. Streptococcus pyogenes (including streptococcal toxic shock syndrome and necrotizing fasciitis). In: Mandell GL, Douglas RG, Dolin R, editors. Principles and practice of infectious diseases, 5th ed., vol. 2. Philadelphia: Churchill Livingstone; 2000. p. 2101–17.
- Ralph AP, Carapetis JR. Group a streptococcal diseases and their global burden. Curr Top Microbiol Immunol. 2013;368:1–27.PubMedGoogle Scholar
- Walker MJ, Barnett TC, McArthur JD, Cole JN, Gillen CM, Henningham A, et al. Disease manifestations and pathogenic mechanisms of Group A Streptococcus. Clin Microbiol Rev. 2014;27:264–301. DOIPubMedGoogle Scholar
- Bessen DE. Population biology of the human restricted pathogen, Streptococcus pyogenes. Infect Genet Evol. 2009;9:581–93. DOIPubMedGoogle Scholar
- Franklinos LH, Efstratiou A, Macgregor SK, John SK, Hopkins T, Cunningham AA, et al. Streptococcus pyogenes infection in a free-living European hedgehog (Erinaceus europaeus). EcoHealth. 2015;12:689–92. DOIPubMedGoogle Scholar
- Willard MD, Berridge B, Braniecki A, Bouley D. Possible antibiotic-associated colitis in a dog. J Am Vet Med Assoc. 1998;213:1775–9, 1753–4.PubMedGoogle Scholar
- Sprot H, Efstratiou A, Hubble M, Morgan M. Man’s best friend?—first report of prosthetic joint infection with Streptococcus pyogenes from a canine source. J Infect. 2012;64:625–7. DOIPubMedGoogle Scholar
- Vela AI, Fernández E, Lawson PA, Latre MV, Falsen E, Domínguez L, et al. Streptococcus entericus sp. nov., isolated from cattle intestine. Int J Syst Evol Microbiol. 2002;52:665–9. DOIPubMedGoogle Scholar
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; twenty-second informational supplement (M100–S22). Wayne (PA): The Institute; 2012.
- Comité de l’antibiogramme de la Societé Française de Microbiologie. Recomendations 2012. Paris: Societé Française de Microbiologie; 2012 [cited 2015 Jul 8]. http://www.sfm-microbiologie.org/
- Hasenbein ME, Warner JE, Lambert KG, Cole SE, Onderdonk AB, McAdam AJ. Detection of multiple macrolide- and lincosamide-resistant strains of Streptococcus pyogenes from patients in the Boston area. J Clin Microbiol. 2004;42:1559–63. DOIPubMedGoogle Scholar
- Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L. Detection of erythromycin-resistant determinants by PCR. Antimicrob Agents Chemother. 1996;40:2562–6.PubMedGoogle Scholar
- Seppälä H, Skurnik M, Soini H, Roberts MC, Huovinen P. A novel erythromycin resistance methylase gene (ermTR) in Streptococcus pyogenes. Antimicrob Agents Chemother. 1998;42:257–62.PubMedGoogle Scholar
- Malhotra-Kumar S, Lammens C, Piessens J, Goossens H. Multiplex PCR for simultaneous detection of macrolide and tetracycline resistance determinants in streptococci. Antimicrob Agents Chemother. 2005;49:4798–800. DOIPubMedGoogle Scholar
- Lüthje P, Schwarz S. Molecular basis of resistance to macrolides and lincosamides among staphylococci and streptococci from various animal sources collected in the resistance monitoring program BfT-GermVet. Int J Antimicrob Agents. 2007;29:528–35. DOIPubMedGoogle Scholar
- Commons R, Rogers S, Gooding T, Danchin M, Carapetis J, Robins-Browne R, et al. Superantigen genes in group A streptococcal isolates and their relationship with emm types. J Med Microbiol. 2008;57:1238–46. DOIPubMedGoogle Scholar
- Rivera A, Rebollo M, Miró E, Mateo M, Navarro F, Gurguí M, et al. Superantigen gene profile, emm type and antibiotic resistance genes among group A streptococcal isolates from Barcelona, Spain. J Med Microbiol. 2006;55:1115–23. DOIPubMedGoogle Scholar
- Stanley J, Linton D, Desai M, Efstratiou A, George R. Molecular subtyping of prevalent M serotypes of Streptococcus pyogenes causing invasive disease. J Clin Microbiol. 1995;33:2850–5.PubMedGoogle Scholar
- Enright MC, Spratt BG, Kalia A, Cross JH, Bessen DE. Multilocus sequence typing of Streptococcus pyogenes and the relationships between emm type and clone. Infect Immun. 2001;69:2416–27. DOIPubMedGoogle Scholar
- Foster AP. Staphylococcal skin disease in livestock. Vet Dermatol. 2012;23:342–51, e63. DOIPubMedGoogle Scholar
- Enright MC, Spratt BG, Kalia A, Cross JH, Bessen DE. Multilocus sequence typing of Streptococcus pyogenes and the relationships between emm type and clone. Infect Immun. 2001;69:2416–27. DOIPubMedGoogle Scholar
- Steer AC, Law I, Matatolu L, Beall BW, Carapetis JR. Global emm type distribution of group A streptococci: systematic review and implications for vaccine development. Lancet Infect Dis. 2009;9:611–6. DOIPubMedGoogle Scholar
- Luca-Harari B, Ekelund K, van der Linden M, Staum-Kaltoft M, Hammerum AM, Jasir A. Clinical and epidemiological aspects of invasive Streptococcus pyogenes infections in Denmark during 2003 and 2004. J Clin Microbiol. 2008;46:79–86. DOIPubMedGoogle Scholar
- Silva-Costa C, Pinto FR, Ramírez M, Melo-Cristino J; Portuguese Surveillance Group for the Study of Respiratory Pathogens. Decrease in macrolide resistance and clonal instability among Streptococcus pyogenes in Portugal. Clin Microbiol Infect. 2008;14:1152–9. DOIPubMedGoogle Scholar
- McGregor KF, Spratt BG. Identity and prevalence of multilocus sequence typing-defined clones of group A streptococci within a hospital setting. J Clin Microbiol. 2005;43:1963–7. DOIPubMedGoogle Scholar
- Reinert RR, Lütticken R, Sutcliffe JA, Tait-Kamradt A, Cil MY, Schorn HM, et al. Clonal relatedness of erythromycin-resistant Streptococcus pyogenes isolates in Germany. Antimicrob Agents Chemother. 2004;48:1369–73. DOIPubMedGoogle Scholar
- Szczypa K, Sadowy E, Izdebski R, Hryniewicz W. A rapid increase in macrolide resistance in Streptococcus pyogenes isolated in Poland during 1996-2002. J Antimicrob Chemother. 2004;54:828–31. DOIPubMedGoogle Scholar
- Ardanuy C, Domenech A, Rolo D, Calatayud L, Tubau F, Ayats J, et al. Molecular characterization of macrolide- and multidrug-resistant Streptococcus pyogenes isolated from adult patients in Barcelona, Spain (1993-2008). J Antimicrob Chemother. 2010;65:634–43. DOIPubMedGoogle Scholar
- Rubio-López V, Valdezate S, Alvarez D, Villalón P, Medina MJ, Salcedo C, et al. Molecular epidemiology, antimicrobial susceptibilities and resistance mechanisms of Streptococcus pyogenes isolates resistant to erythromycin and tetracycline in Spain (1994-2006). BMC Microbiol. 2012;12:215. DOIPubMedGoogle Scholar
- Rivera A, Rebollo M, Miró E, Mateo M, Navarro F, Gurguí M, et al. Superantigen gene profile, emm type and antibiotic resistance genes among group A streptococcal isolates from Barcelona, Spain. J Med Microbiol. 2006;55:1115–23. DOIPubMedGoogle Scholar
- Opavski N, Gajic I, Borek AL, Obszańska K, Stanojevic M, Lazarevic I, et al. Molecular characterization of macrolide resistant Streptococcus pyogenes isolates from pharyngitis patients in Serbia. Infect Genet Evol. 2015;33:246–52. DOIPubMedGoogle Scholar
- Lin JN, Chang LL, Lai CH, Lin HH, Chen YH. Clinical and molecular characteristics of invasive and noninvasive skin and soft tissue infections caused by group A Streptococcus. J Clin Microbiol. 2011;49:3632–7. DOIPubMedGoogle Scholar
- Pérez-Trallero E, Montes M, Orden B, Tamayo E, García-Arenzana JM, Marimón JM. Phenotypic and genotypic characterization of Streptococcus pyogenes isolates displaying the MLSB phenotype of macrolide resistance in Spain, 1999 to 2005. Antimicrob Agents Chemother. 2007;51:1228–33. DOIPubMedGoogle Scholar
- Sørensen UB, Poulsen K, Ghezzo C, Margarit I, Kilian M. Emergence and global dissemination of host-specific Streptococcus agalactiae clones. MBio. 2010;1:e00178–10. DOIPubMedGoogle Scholar
- Jordan HT, Richards CL Jr, Burton DC, Thigpen MC, Van Beneden CA. Group a streptococcal disease in long-term care facilities: descriptive epidemiology and potential control measures. Clin Infect Dis. 2007;45:742–52. DOIPubMedGoogle Scholar
- Acikgoz ZC, Gocer S, Tuncer S. Macrolide resistance determinants of group A streptococci in Ankara, Turkey. J Antimicrob Chemother. 2003;52:110–2. DOIPubMedGoogle Scholar
- Szczypa K, Sadowy E, Izdebski R, Hryniewicz W. A rapid increase in macrolide resistance in Streptococcus pyogenes isolated in Poland during 1996-2002. J Antimicrob Chemother. 2004;54:828–31. DOIPubMedGoogle Scholar
- Richter SS, Heilmann KP, Beekmann SE, Miller NJ, Miller AL, Rice CL, et al. Macrolide-resistant Streptococcus pyogenes in the United States, 2002-2003. Clin Infect Dis. 2005;41:599–608. DOIPubMedGoogle Scholar
- Pavlovic L, Grego E, Sipetic-Grujicic S. Prevalence of macrolide resistance in Streptococcus pyogenes collected in Serbia. Jpn J Infect Dis. 2010;63:275–6.PubMedGoogle Scholar
- Brenciani A, Bacciaglia A, Vecchi M, Vitali LA, Varaldo PE, Giovanetti E. Genetic elements carrying erm(B) in Streptococcus pyogenes and association with tet(M) tetracycline resistance gene. Antimicrob Agents Chemother. 2007;51:1209–16. DOIPubMedGoogle Scholar
- Giovanetti E, Montanari MP, Mingoia M, Varaldo PE. Phenotypes and genotypes of erythromycin-resistant Streptococcus pyogenes strains in Italy and heterogeneity of inducibly resistant strains. Antimicrob Agents Chemother. 1999;43:1935–40.PubMedGoogle Scholar
- Giovanetti E, Brenciani A, Lupidi R, Roberts MC, Varaldo PE. Presence of the tet(O) gene in erythromycin- and tetracycline-resistant strains of Streptococcus pyogenes and linkage with either the mef(A) or the erm(A) gene. Antimicrob Agents Chemother. 2003;47:2844–9. DOIPubMedGoogle Scholar
- Dundar D, Sayan M, Tamer GS. Macrolide and tetracycline resistance and emm type distribution of Streptococcus pyogenes isolates recovered from Turkish patients. Microb Drug Resist. 2010;16:279–84.
Figure
Tables
Cite This ArticleTable of Contents – Volume 23, Number 12—December 2017
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Jose F. Fernández-Garayzábal, Universidad Complutense de Madrid, Ftad. de Veterinaria–Patologia Animal I (Sanidad Animal), Avda. Puerta de Hierro s/n n/a, Madrid, Madrid 28040 Spain
Top