Volume 23, Number 12—December 2017
Research
High Rate of MCR-1–Producing Escherichia coli and Klebsiella pneumoniae among Pigs, Portugal
Cite This Article
Citation for Media
Abstract
The mcr-1 (mobile colistin resistance 1) gene, which encodes phosphoethanolamine transferase, has been recently identified as a source of acquired resistance to polymyxins in Escherichia coli. Using the SuperPolymyxin selective medium, we prospectively screened 100 pigs at 2 farms in Portugal for polymyxin-resistant Enterobacteriaceae and recovered 98 plasmid-mediated MCR-1–producing isolates. Most isolates corresponded to nonclonally related E. coli belonging to many sequence types; we also found 2 Klebsiella pneumoniae sequence types. The mcr-1 gene was carried on IncHI2 or IncP plasmid backbones. Our finding of a high rate of MCR-1 producers on 2 pig farms in Portugal highlights the diffusion of that colistin-resistance determinant at the farm level. The fact that the pigs received colistin as metaphylaxis in their feed during the 6 weeks before sampling suggests selective pressure.
The progressive global increase of antimicrobial drug resistance in Enterobacteriaceae is worrisome, and adding to the concern is the recent discovery of the plasmid-mediated mobile colistin resistance (MCR) genes mcr-1 and mcr-2 (1,2). These genes encode phosphoethanolamine transferases, which add a phosphoethanolamine group to the lipid A of the lipopolysaccharide, leading to gram-negative bacteria resistance to polymyxins (3). Since its discovery, the mcr-1 gene has been identified almost worldwide, mostly in animal and environmental samples (3) and to a lesser extent in human clinical samples (4). The mcr-1 gene has often been identified from Escherichia coli strains recovered from pigs (3,5–8). More recently, the mcr-2 gene, which shares 76.8% nt identity with mcr-1, has been identified from a single E. coli isolate recovered from a pig in Belgium (2). The genetic element related to the mcr-2 gene and possibly involved in its acquisition is insertion sequence (IS) ISEc69.
The mcr-1 gene has been identified on a large variety of plasmids, such as IncI2, IncX4, IncHI2, IncP, IncFI, IncFII, IncFIB, and IncY (3,9,10). The genetic context of the mcr-1 gene always includes the mcr-1 cassette, as previously described (11,12). In addition, ISApl1 is often found upstream of the mcr-1 gene. It has been recently shown that a second copy of ISApl1 may be found downstream of the mcr-1 gene, therefore bracketing the 2.6-kb mcr-1 cassette and forming the composite transposon Tn6330, demonstrated to be functional and responsible for the transposition of mcr-1 (13,14).
We speculate that the emergence and further dissemination of the mcr-1 and -2 genes occurred from pigs and that ISApl and ISEc69, respectively, were the main genetic elements involved in that process. We recently demonstrated that Moraxella spp. are sources of mcr-like encoding genes (15); M. pluranimalium has been identified as the progenitor of the mcr-2 gene (16). Of note, all Moraxella spp. are widespread in pigs (17), thus allowing speculation that the whole genetic process that originally led to the emergence of the mcr-like genes occurred in those animals.
Worldwide, colistin is widely used in veterinary medicine for different purposes, including treatment of enteric infections, prophylaxis or metaphylaxis (18), and as growth promoter in several countries (19). Despite this selective pressure, studies reporting identification of colistin-resistant Enterobacteriaceae in veterinary medicine remain scarce, although an overall low prevalence of those resistant strains was noticed in Europe (8,10,20,21).
To evaluate the prevalence and genetic characterization of colistin-resistant Enterobacteriaceae on pig farms, we performed a prospective epidemiologic survey. The study was conducted in Portugal (the fifth largest polymyxin consumer in Europe), where colistin is heavily used in veterinary medicine (22).
Isolates and Susceptibility Testing
On 1 day in June 2016, we collected 100 rectal swab samples from pigs on 2 pig farms in Portugal, 30 km apart. These farms, harboring ≈3,000 pigs each, are production holdings, where piglets are born and fattened before being delivered to slaughterhouses (23). All pigs sampled were 10–11 weeks of age. When the pigs were 5–10 weeks of age, their feed included colistin (0.5%), amoxicillin (0.5%), and zinc oxide (0.15%). The weekly dose of colistin in the regimen was ≈0.06 g/kg. Overall, all pigs received ≈5.5 g colistin for metaphylaxis over 6 weeks.
We incubated rectal swab samples overnight at 37°C in Luria-Bertani broth supplemented with 1 μg/mL colistin. The next day, to select for colistin-resistant gram-negative isolates, we inoculated each enrichment tube onto SuperPolymyxin selective agar medium that contained 3.5 μg/mL colistin and 10 μg/mL daptomycin (ELITechGroup, Signes, France) (24). We identified colistin-resistant isolates recovered from SuperPolymyxin plates with API 20E (bioMérieux, La Balme les Grottes, France). We performed antimicrobial drug susceptibility testing by using the disk-diffusion method according to Clinical and Laboratory Standards Institute recommendations, on Muller-Hinton agar plates, except for colistin, for which we evaluated MICs for colistin by broth microdilution in cation-adjusted Muller-Hinton broth (Bio-Rad, Cressier, Switzerland), as recommended by the Clinical and Laboratory Standards Institute (25).
Molecular Analyses
Acquired colistin-resistance genes mcr-1 and mcr-2 were identified by PCR, with use of specific primers as reported (14), and amplicons were further sequenced by Microsynth (Balgach, Switzerland). We identified extended-spectrum β-lactamase (ESBL)–encoding genes by using primers specific for detection of blaTEM, blaSHV, and blaCTX-M genes (26,27). The plasmidborne chloramphenicol gene floR was sought by using specific primers (28) among isolates exhibiting resistance to chloramphenicol. The clonal relationship of the colistin-resistant isolates was evaluated by pulsed-field gel electrophoresis, as described previously (29), and multilocus sequence typing was performed for a representative strain of each pulsotype. We assigned sequence types (STs) by using the multilocus sequence typing databases for E. coli (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli) and K. pneumoniae (http://bigsdb.pasteur.fr/klebsiella/klebsiella.html). The phylogenetic group of E. coli isolates was determined with the PCR-based Clermont method as described previously (30).
Conjugation Experiments and Plasmid Analyses
We performed conjugation assays on filters with azide-resistant E. coli J53 as the recipient strain. MCR-1 producers and the J53 isolate were cultured overnight in Luria-Bertani broth. To reach the logarithmic phase, the donor and recipient strains were subcultured in fresh Luria-Bertani broth for 3 h. We subsequently mixed the samples at a ratio of 10:1 and deposited 100 μL of this mix onto 22-μm filters, which we then incubated for 5 h at 37°C on Luria-Bertani agar plates. After the incubation, filters were resuspended in 0.85% NaCl, and we plated 250 μL of this mixture onto selective Luria-Bertani plates containing azide (100 μg/mL) and colistin (1 μg/mL). PCR was used to check all E. coli transconjugants for the mcr-1 gene.
We typed plasmids carrying the mcr-1 gene from E. coli transconjugants by using the PCR-based replicon typing method (31). The size of the plasmid was obtained after Kieser extraction (32) and agarose gel electrophoresis; we used as a reference E. coli 50192 isolate containing 4 characterized plasmids (154 kb, 66 kb, 48 kb, and 7 kb).
From the 100 rectal swab samples collected, we recovered 108 colistin-resistant isolates from the SuperPolymyxin agar plates and identified 90 as E. coli, 17 as K. pneumoniae, and 1 as Proteus mirabilis. Of the 108 colistin-resistant isolates, 98 were positive for the mcr-1 gene. Colistin MICs for all MCR-1–producing isolates ranged from 4 to 32 μg/mL (Table). Among these positive isolates, 10 showed an ESBL phenotype. Sequencing revealed that all mcr-positive isolates possessed a gene that was 100% identical to mcr-1. All MCR-1–producing isolates possessed the blaTEM-1 gene, and all ESBL producers possessed the blaCTX-M-2 gene. All MCR-1–producing isolates were resistant to penicillins and tetracycline, 97.9% were resistant to sulfamethoxazole/trimethoprim, 96% were resistant to tobramycin, and 84.7% were resistant to chloramphenicol. Among the chloramphenicol-resistant isolates, 56% were positive for the floR resistance gene. No mcr-2–positive isolate was identified in our samples.
Pulsed-field gel electrophoresis identified 19 distinct E. coli clones from the first farm sampled and 18 from the second (Table). The isolates belonged to 15 STs (ST10, ST23, ST38, ST46, ST101, ST156, ST6453, and 8 new STs); only 2 (ST10 and ST101) were detected on both farms. Phylogenetic typing showed that each E. coli isolate belonged to 1 of the phylogroups A, B1, C, D, E, or F. No extraintestinal and virulent B2 phylogroup was detected among all E. coli isolates. In addition, we identified 2 K. pneumoniae STs (1 clone per farm), ST45 and ST1563.
Conjugation followed by PCR-based replicon typing analysis showed that the mcr-1 gene was carried on different plasmids (Table). The mcr-1 gene was identified on IncHI2 (54%), IncP (38%), and IncX4 (8%) plasmids. Conjugation experiments showed that resistance to sulfamethoxazole/trimethoprim and sulfonamides was systematically co-transferred along with the mcr-1 gene when carried by IncHI2 plasmids and that resistance to tetracycline, tobramycin, chloramphenicol, and amoxicillin was also most often co-transferred by IncP plasmids (Table). Conversely, the mcr-1 gene was the only resistance determinant when located on IncX4 plasmids. Of note, IncP-type plasmids carrying mcr-1 were predominant on the first farm, whereas IncHI2 plasmids were predominant on the second farm.
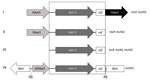
By PCR mapping using previously published primers (12,13), in all isolates we identified the ≈2.6-kb mcr-1 cassette in association with 0, 1, or 2 copies of insertion sequence ISApl1, depending on the isolates tested (Table; Figure). In addition, we found a genetic structure not previously reported in 1 isolate that consisted of a truncated ISApl1 element upstream of the mcr-1 cassette. That structure, inserted into a kinase gene onto an IncHI2 plasmid, corresponded to a truncated version of transposon Tn6330 previously reported, with a 2-bp AG direct repeat bracketing the ∆ISApl1-mcr-1 structure, suggesting a former insertion event through a transposition mechanism (Figure).
Prevalence of MCR-1–producing Enterobacteriacae isolates, mainly E. coli, from 2 pig farms in Portugal was high. So far, the mcr-1 gene has been identified mainly in animal samples, rarely in environmental and human samples (3,4,33–36). Previous studies describe the occurrence of MCR-1 producers in swine; prevalence in Europe ranged from 0.5% to 13.5% (6,8,36,37) and in China up to 20.6% (1). In our study, we detected an extremely high rate of MCR-1–producing Enterobacteriaceae, finding the mcr-1 gene in isolates from 98% of pigs tested. We identified an MCR-1–producing K. pneumoniae among the pigs, which is noteworthy considering the infrequent recovery of K. pneumoniae from animals. Nevertheless, most of the mcr-1–positive isolates identified here were E. coli, as is reported in most epidemiologic studies (3). For the same pigs that had been screened (by nasal swabs) for methicillin-resistant Staphylococcus aureus, the rate of colonization was very high (99%); 2 main spa types of clone ST398 were identified (23).
Our study was performed with samples from pigs, and it would be of interest to conduct similar studies of humans. In France, a survey performed in a hospital during February–May 2016 (38) showed a high rate (23%) of fecal carriage with intrinsic colistin-resistant gram-negative isolates but a low rate (1.4%) of acquired polymyxin resistance; no mcr-1 or mcr-2 genes were identified. A retrospective study focusing on Salmonella isolates was previously performed in Portugal, and MCR-1 producers were reportedly found in humans and pork (39). No MCR-2–producing isolate was identified in our study, although this gene was also identified in pigs (2).
We used the newly developed SuperPolymyxin medium for our prospective epidemiologic study. The fact that no colistin-susceptible strain was recovered during the screening further highlights the excellent specificity of this medium.
Unexpectedly, we found that the studied collection of MCR-1 producers was highly diverse; we identified many STs and genetic features associated with mcr-1. The rate of mcr-1 in our study was very high and caused by the dissemination of neither a single clone nor a plasmid. This high diversity could be explained by 2 key elements. First, considering that the selective pre-enrichment with Luria-Bertani broth supplemented with 1 μg/mL of colistin and subsequent culturing on the Superpolymyxin medium exhibits a high sensitivity, such a prospective survey may detect higher rates of colistin-resistant isolates than would previous studies. Second, the presence of colistin in the pig food on the 2 farms studied probably represents an efficient selective pressure for MCR-1 acquisition. Findings of similar and comparative studies performed in countries that do not use polymyxins in animals, such as Norway or Finland, would be of interest.
Several STs of E. coli identified in this study were similar to those of other MCR-1– producing isolates reported from other studies. As an example, ST10 E. coli producing MCR-1 was recovered from swine farms in Germany and in clinical samples in India and South Africa (6,11,40). In South Africa, ST101 E. coli was identified from a patient with a urinary tract infection (11). ST156 E. coli was identified at a hospital in China and in a muscovy duck in China, where it was co-producing MCR-1 and NDM (New Delhi metallo-β-lactamase)–5 carbapenemase (41,42). We showed that all E. coli isolates belonged to a commensal population and not to extraintestinal pathogenic strains, which is consistent with findings of other studies (3) and in line with the design of our study, which was analyzing the colonizing gut flora.
We showed that the mcr-1 gene was carried by a diversity of plasmids. However, most plasmids recovered from the first farm were ≈60-kb IncP plasmids, whereas those from the second farm were ≈250-kb IncHI2 plasmids. Only 3 STs possessed a ≈30-kb IncX4 plasmid carrying the mcr-1 gene, which contrasts with other studies that have shown this type of plasmid to be predominant (43). In accordance with what has been observed with other characterized IncX4 plasmids, we found no other resistant determinant associated with mcr-1 on that plasmid type. IncP and IncHI2 plasmids carried other resistance determinants associated with mcr-1. Remarkably, we found no IncI2-type plasmid carrying mcr-1 in those isolates, although they have often been reported in the literature (1,3).
Analysis of the genetic features associated with the mcr-1 gene further highlights that it was probably originally acquired by a transposition mechanism and that ISApl1 played a major role; further truncations or rearrangements led to the stabilization of this structure, as suggested by Snesrud et al. (44). We also identified the entire composite transposon Tn6330 comprising 2 copies of ISApl1 bracketing the mcr-1 cassette (13). Because this entire transposon was detected in some isolates of this collection in addition to other defective versions of it, we can speculate that this structure may still be mobilizable and continue to disseminate between different genetic locations.
In summary, the rate of pig colonization with MCR-1–producing Enterobacteriaceae was high at the 2 farms we sampled, showing substantial diversity of species, clonality, and genetic aspects. Even if these results suggest that colistin constitutes a major driving force for selecting plasmids carrying the mcr-1 gene, the occurrence of the blaTEM-1 gene on the same plasmid indicates that β-lactams might also be co-selecting for colistin resistance through the acquisition of such plasmids. In addition, this study showed that SuperPolymyxin is an efficient medium for screening colistin-resistant isolates from animal samples and performing such epidemiologic surveys. Last, considering that a recent report from Germany identified pig farms as potential sources of environmental contamination for MCR-1–producing E. coli (6), our data strongly indicate the need for screening farm environments in Portugal, to evaluate the extent to which the spread of those resistant bacteria has already occurred and, therefore, to better measure the risk to human health.
Mr. Kieffer is a PhD student in microbiology at the University of Fribourg, Switzerland. His main activities are related to the identification of acquired polymyxin and carbapenem resistance mechanisms in gram-negative bacteria, with a focus on the genetics of resistance gene acquisition.
Acknowledgments
We are grateful to S. Kumar-Malhotra for the gift of the MCR-2–producing E. coli strain.
This work has been funded by the University of Fribourg; by grants from the Animal Health and Welfare ERA-Net project, Switzerland; by the Office Fédéral de la Santé Publique, Bern, Switzerland (grant no. 16009294); by project PTDC/DTP-EPI/0842/2014 from Fundação para a Ciência e a Tecnologia, Portugal; and by the Novartis Foundation for Medical-Biological Research.
An international patent form has been filed on behalf of the University of Fribourg (Switzerland) corresponding to the SuperPolymyxin selective medium.
References
- Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16:161–8. DOIPubMedGoogle Scholar
- Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill. 2016;21:30280. DOIPubMedGoogle Scholar
- Poirel L, Jayol A, Nordmann P. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev. 2017;30:557–96. DOIPubMedGoogle Scholar
- Liassine N, Assouvie L, Descombes MC, Tendon VD, Kieffer N, Poirel L, et al. Very low prevalence of MCR-1/MCR-2 plasmid-mediated colistin resistance in urinary tract Enterobacteriaceae in Switzerland. Int J Infect Dis. 2016;51:4–5. DOIPubMedGoogle Scholar
- Irrgang A, Roschanski N, Tenhagen BA, Grobbel M, Skladnikiewicz-Ziemer T, Thomas K, et al. Prevalence of mcr-1 in E. coli from livestock and food in Germany, 2010–2015. PLoS One. 2016;11:e0159863. DOIPubMedGoogle Scholar
- Guenther S, Falgenhauer L, Semmler T, Imirzalioglu C, Chakraborty T, Roesler U, et al. Environmental emission of multiresistant Escherichia coli carrying the colistin resistance gene mcr-1 from German swine farms. J Antimicrob Chemother. 2017;72:1289–92.PubMedGoogle Scholar
- Huang X, Yu L, Chen X, Zhi C, Yao X, Liu Y, et al. High prevalence of colistin resistance and mcr-1 gene in Escherichia coli isolated from food animals in China. Front Microbiol. 2017;8:562. DOIPubMedGoogle Scholar
- Perrin-Guyomard A, Bruneau M, Houée P, Deleurme K, Legrandois P, Poirier C, et al. Prevalence of mcr-1 in commensal Escherichia coli from French livestock, 2007 to 2014. Euro Surveill. 2016;21:30135. DOIPubMedGoogle Scholar
- Zhang C, Feng Y, Liu F, Jiang H, Qu Z, Lei M, et al. A phage-like IncY plasmid carrying the mcr-1 gene in Escherichia coli from a pig farm in China. Antimicrob Agents Chemother. 2017;61:e02035–16. DOIPubMedGoogle Scholar
- Xavier BB, Lammens C, Butaye P, Goossens H, Malhotra-Kumar S. Complete sequence of an IncFII plasmid harbouring the colistin resistance gene mcr-1 isolated from Belgian pig farms. J Antimicrob Chemother. 2016;71:2342–4. DOIPubMedGoogle Scholar
- Poirel L, Kieffer N, Brink A, Coetze J, Jayol A, Nordmann P. Genetic features of MCR-1–producing colistin-resistant Escherichia coli isolates in South Africa. Antimicrob Agents Chemother. 2016;60:4394–7. DOIPubMedGoogle Scholar
- Zurfluh K, Kieffer N, Poirel L, Nordmann P, Stephan R. Features of the mcr-1 cassette related to colistin resistance. Antimicrob Agents Chemother. 2016;60:6438–9. DOIPubMedGoogle Scholar
- Poirel L, Kieffer N, Nordmann P. In-vitro study of ISApl1-mediated mobilization of the colistin resistance gene mcr-1. Antimicrob Agents Chemother. 2017;61:e00127–17. DOIPubMedGoogle Scholar
- Zurfluh K, Klumpp J, Nüesch-Inderbinen M, Stephan R. Full-length nucleotide sequences of mcr-1–harboring plasmids isolated from extended-spectrum-β-lactamase-producing Escherichia coli isolates of different origins. Antimicrob Agents Chemother. 2016;60:5589–91. DOIPubMedGoogle Scholar
- Kieffer N, Nordmann P, Poirel L. Moraxella species as potential sources of MCR-like polymyxin-resistance determinants. Antimicrob Agents Chemother. 2017;61:e00129–17. DOIPubMedGoogle Scholar
- Poirel L, Kieffer N, Fernandez-Garayzabal JF, Vela AI, Larpin Y, Nordmann P. MCR-2–mediated plasmid-borne polymyxin resistance most likely originates from Moraxella pluranimalium. J Antimicrob Chemother. 2017;72:2947–9. DOIGoogle Scholar
- Vela AI, Sánchez-Porro C, Aragón V, Olvera A, Domínguez L, Ventosa A, et al. Moraxella porci sp. nov., isolated from pigs. Int J Syst Evol Microbiol. 2010;60:2446–50. DOIPubMedGoogle Scholar
- Rhouma M, Beaudry F, Thériault W, Letellier A. Colistin in pig production: chemistry, mechanism of antibacterial action, microbial resistance emergence, and One Health perspectives. Front Microbiol. 2016;7:1789. DOIPubMedGoogle Scholar
- Katsunuma Y, Hanazumi M, Fujisaki H, Minato H, Hashimoto Y, Yonemochi C. Associations between the use of antimicrobial agents for growth promotion and the occurrence of antimicrobial-resistant Escherichia coli and enterococci in the feces of livestock and livestock farmers in Japan. J Gen Appl Microbiol. 2007;53:273–9. DOIPubMedGoogle Scholar
- Quesada A, Ugarte-Ruiz M, Iglesias MR, Porrero MC, Martínez R, Florez-Cuadrado D, et al. Detection of plasmid mediated colistin resistance (MCR-1) in Escherichia coli and Salmonella enterica isolated from poultry and swine in Spain. Res Vet Sci. 2016;105:134–5. DOIPubMedGoogle Scholar
- Falgenhauer L, Waezsada SE, Yao Y, Imirzalioglu C, Käsbohrer A, Roesler U, et al.; RESET consortium. Colistin resistance gene mcr-1 in extended-spectrum β-lactamase-producing and carbapenemase-producing Gram-negative bacteria in Germany. Lancet Infect Dis. 2016;16:282–3. DOIPubMedGoogle Scholar
- Catry B, Cavaleri M, Baptiste K, Grave K, Grein K, Holm A, et al. Use of colistin-containing products within the European Union and European Economic Area (EU/EEA): development of resistance in animals and possible impact on human and animal health. Int J Antimicrob Agents. 2015;46:297–306. DOIPubMedGoogle Scholar
- Conceição T, de Lencastre H, Aires-de-Sousa M. Frequent isolation of methicillin resistant Staphylococcus aureus (MRSA) ST398 among healthy pigs in Portugal. PLoS One. 2017;12:e0175340. DOIPubMedGoogle Scholar
- Nordmann P, Jayol A, Poirel L. A universal culture medium for screening polymyxin-resistant Gram-negative isolates. J Clin Microbiol. 2016;54:1395–9. DOIPubMedGoogle Scholar
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; 26th informational supplement (M100–S26). Wayne (PA): The Institute; 2016.
- Lartigue MF, Poirel L, Nordmann P. Diversity of genetic environment of bla(CTX-M) genes. FEMS Microbiol Lett. 2004;234:201–7. DOIPubMedGoogle Scholar
- Kim J, Lim YM. Prevalence of derepressed ampC mutants and extended-spectrum β-lactamase producers among clinical isolates of Citrobacter freundii, Enterobacter spp., and Serratia marcescens in Korea: dissemination of CTX-M-3, TEM-52, and SHV-12. J Clin Microbiol. 2005;43:2452–5. DOIPubMedGoogle Scholar
- Poirel L, Kieffer N, Liassine N, Thanh D, Nordmann P. Plasmid-mediated carbapenem and colistin resistance in a clinical isolate of Escherichia coli. Lancet Infect Dis. 2016;16:281. DOIPubMedGoogle Scholar
- Kieffer N, Nordmann P, Aires-de-Sousa M, Poirel L. High prevalence of carbapenemase-producing Enterobacteriaceae among hospitalized children in Luanda, Angola. Antimicrob Agents Chemother. 2016;60:6189–92. DOIPubMedGoogle Scholar
- Clermont O, Christenson JK, Denamur E, Gordon DM. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep. 2013;5:58–65. DOIPubMedGoogle Scholar
- Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63:219–28. DOIPubMedGoogle Scholar
- Kieser T. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid. 1984;12:19–36. DOIPubMedGoogle Scholar
- Nordmann P, Assouvie L, Prod’Hom G, Poirel L, Greub G. Screening of plasmid-mediated MCR-1 colistin-resistance from bacteremia. Eur J Clin Microbiol Infect Dis. 2016;35:1891–2. DOIPubMedGoogle Scholar
- Jiang X, Poirel L, Nordmann P. Lack of polymyxin resistance among carbapenemase-producing Enterobacteriaceae in a university hospital in China. Infect Dis (Lond). 2017;49:556–7. DOIPubMedGoogle Scholar
- Buess S, Nüesch-Inderbinen M, Stephan R, Zurfluh K. Assessment of animals as a reservoir for colistin resistance: No MCR-1/MCR-2-producing Enterobacteriaceae detected in Swiss livestock. J Glob Antimicrob Resist. 2017;8:33–4. DOIPubMedGoogle Scholar
- Malhotra-Kumar S, Xavier BB, Das AJ, Lammens C, Butaye P, Goossens H. Colistin resistance gene mcr-1 harboured on a multidrug resistant plasmid. Lancet Infect Dis. 2016;16:283–4. DOIPubMedGoogle Scholar
- Falgenhauer L, Waezsada SE, Gwozdzinski K, Ghosh H, Doijad S, Bunk B, et al. Chromosomal Locations of mcr-1 and bla CTX-M-15 in Fluoroquinolone-Resistant Escherichia coli ST410. Emerg Infect Dis. 2016;22:1689–91. DOIPubMedGoogle Scholar
- Saly M, Jayol A, Poirel L, Mégraud F, Nordmann P, Dubois V. Prevalence of faecal carriage of colistin-resistant Gram-negative rods in a university hospital in western France, 2016. J Med Microbiol. 2017;66:842–3. DOIPubMedGoogle Scholar
- Campos J, Cristino L, Peixe L, Antunes P. MCR-1 in multidrug-resistant and copper-tolerant clinically relevant Salmonella 1,4,[5],12:i:- and S. Rissen clones in Portugal, 2011 to 2015. Euro Surveill. 2016;21:30270. DOIPubMedGoogle Scholar
- Bernasconi OJ, Kuenzli E, Pires J, Tinguely R, Carattoli A, Hatz C, et al. Travelers can import colistin-resistant Enterobacteriaceae, including those possessing the plasmid-mediated mcr-1 gene. Antimicrob Agents Chemother. 2016;60:5080–4. DOIPubMedGoogle Scholar
- Yang RS, Feng Y, Lv XY, Duan JH, Chen J, Fang LX, et al. Emergence of NDM-5- and MCR-1-producing Escherichia coli clones ST648 and ST156 from a single muscovy duck (Cairina moschata). Antimicrob Agents Chemother. 2016;60:6899–902. DOIPubMedGoogle Scholar
- He QW, Xu XH, Lan FJ, Zhao ZC, Wu ZY, Cao YP, et al. Molecular characteristic of mcr-1 producing Escherichia coli in a Chinese university hospital. Ann Clin Microbiol Antimicrob. 2017;16:32. DOIPubMedGoogle Scholar
- Sun J, Fang LX, Wu Z, Deng H, Yang RS, Li XP, et al. Genetic analysis of the IncX4 plasmids: implications for a unique pattern in the mcr-1 acquisition. Sci Rep. 2017;7:424. DOIPubMedGoogle Scholar
- Snesrud E, He S, Chandler M, Dekker JP, Hickman AB, McGann P, et al. A model for transposition of the colistin resistance gene mcr-1 by ISApl1. Antimicrob Agents Chemother. 2016;60:6973–6. DOIPubMedGoogle Scholar
Figure
Table
Cite This ArticleTable of Contents – Volume 23, Number 12—December 2017
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Laurent Poirel, Medical and Molecular Microbiology Unit, Department of Medicine, Faculty of Science, University of Fribourg, rue Albert Gockel 3, CH-1700 Fribourg, Switzerland
Top