Volume 23, Number 3—March 2017
Research Letter
Molecular Verification of New World Mansonella perstans Parasitemias
Figure
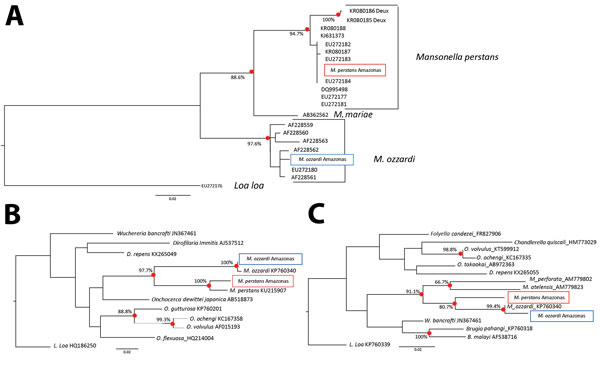
Figure. Maximum-likelihood phylogenetic trees showing the relationship between Mansonella parasites from Amazon region of Brazil (Amazonas state) and some of their closest relatives. A) Ribosomal internal transcribed spacer 1–based phylogeny. B) Mitochondrial cytochrome c oxidase subunit 1–based phylogeny. C) Mitochondrial 12S-based phylogeny. All 3 trees were prepared by using DNA sequence alignments and PHYLIP version 3.67 (http://evolution.genetics.washington.edu/phylip.html). Black circles indicate significant bootstrap-supported nodes as a percentage of 1,000 pseudoreplicates. Solid boxes indicate M. perstans and dashed boxes M. ozzardi sequences generated for this study and used in the construction of the displayed trees. Scale bars indicate nucleotide substitutions per site. These sequence have been submitted to GenBank and EMBL (accession nos.: M. perstans cytochrome c oxidase subunit 1, LT623909; M. ozzardi cytochrome c oxidase subunit 1, LT623910; M. perstans 12S, LT623913; M. ozzardi 12S, LT623914; M. perstans internal transcribed spacer 1, LT623911; and M. ozzardi internal transcribed spacer 1, LT623912).
1These authors contributed equally to this article.