Zoonotic Transmission of mcr-1 Colistin Resistance Gene from Small-Scale Poultry Farms, Vietnam
Nguyen Vinh Trung

, Sébastien Matamoros, Juan J. Carrique-Mas, Nguyen Huu Nghia, Nguyen Thi Nhung, Tran Thi Bich Chieu, Ho Huynh Mai, Willemien van Rooijen, James Campbell, Jaap A. Wagenaar, Anita Hardon, Nguyen Thi Nhu Mai, Thai Quoc Hieu, Guy Thwaites, Menno D. de Jong, Constance Schultsz
1, and Ngo Thi Hoa
1
Author affiliations: University of Amsterdam, Amsterdam, the Netherlands (N.V. Trung, S. Matamoros, W. van Rooijen, A. Hardon, M.D. de Jong, C. Schultsz); Amsterdam Institute for Global Health and Development, Amsterdam (N.V. Trung, S. Matamoros, C. Schultsz); Centre for Tropical Medicine, Ho Chi Minh City, Vietnam (N.V. Trung, J.J. Carrique-Mas, N.H. Nghia, N.T. Nhung, T.T.B. Chieu, J. Campbell, G. Thwaites, C. Schultsz, N.T. Hoa); University of Oxford, Oxford, UK (J.J. Carrique-Mas, J. Campbell, G. Thwaites, N.T. Hoa); Sub-Department of Animal Health, My Tho, Vietnam (H.H. Mai, T.Q. Hieu); Utrecht University, Utrecht, the Netherlands (J.A. Wagenaar); Central Veterinary Institute of Wageningen University & Research, Lelystad, the Netherlands (J.A. Wagenaar); Preventive Medicine Center, My Tho (N.T.N. Mai)
Main Article
Figure
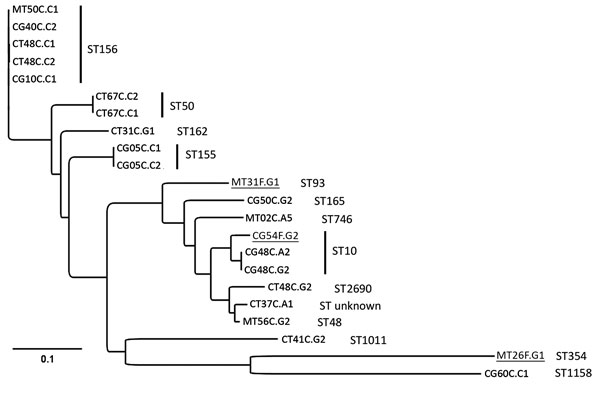
Figure. Phylogenetic analyses of mcr-1–positive Escherichia coli isolated from chickens and chicken farmers, Vietnam, 2012–2013. Maximum-likelihood tree of 22 mcr-1–carrying E. coli isolated from 15 chicken fecal samples and 3 human fecal swab samples (underlined), constructed by using CSI Phylogeny 1.4 (https://cge.cbs.dtu.dk//services/CSIPhylogeny/), shows a genome-wide single-nucleotide polymorphism (SNP) comparison. A total of 74,585 SNPs were concatenated for pairwise comparison (difference between pairs 0–32,267 SNPs). The multilocus sequence types (ST) are indicated next to the isolate names. The ST155 isolates CG05C.C1 and CG05C.C2 differ by 1 SNP; the ST10 isolates CG48C.A2 and CG48C.G2 differ by 1 SNP and 1 antimicrobial resistance gene; the ST156 isolates CT48C.C1 and CT48C.C2 differ by 4 SNPs and 3 antimicrobial resistance genes; and the ST50 isolates CT67C.C1 and CT67C.C2 are phenotypically different but have 0 SNP differences and originate from the same sample and are therefore likely to be highly related or identical. Scale bar indicates number of nucleotide substitutions per site.
Main Article
Page created: February 17, 2017
Page updated: February 17, 2017
Page reviewed: February 17, 2017
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
