Volume 23, Number 4—April 2017
Research
Plasmodium malariae Prevalence and csp Gene Diversity, Kenya, 2014 and 2015
Figure 5
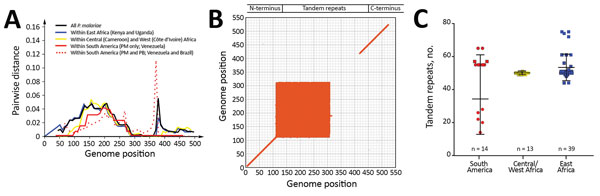
Figure 5. Comparison of circumsporozoite protein (csp) gene sequence divergence among Plasmodium isolates from different geographic regions. A) Pairwise genetic distance plot of all amino acid positions of the csp gene. The matrix-normalized distances based on the standard point accepted mutation (Dayhoff–PAM) model that account for the probability of change from 1 amino acid to another were calculated. Samples were analyzed as a whole and partitioned by geographic regions as indicated by colors. B) Dot plot showing matching scores, a proxy of sequence similarity, between pairwise samples calculated based on the standard Dayhoff–PAM matrix. The greatest mismatch was detected at amino acid positions 110–310, representing the 3′ N terminal through the central repeat regions. C) Variation in the number of tandem repeats in the central region of the csp gene. The greatest length variation was observed in the isolates from South America despite the fact that both P. malariae (PM) and P. brasilianum (PB) were included. P. malariae from East Africa was more variable in the number of repeats than isolates from Central/West Africa, despite difference in sample size. Median, first quartile, and fourth quartile of the data are shown for each sample category (horizontal lines). Red represents samples from South America; yellow, Central/West Africa; blue, East Africa. Circles represent P. brasilianum; squares, P. malariae.