Volume 23, Number 5—May 2017
Research
Population Genomics of Legionella longbeachae and Hidden Complexities of Infection Source Attribution
Figure 3
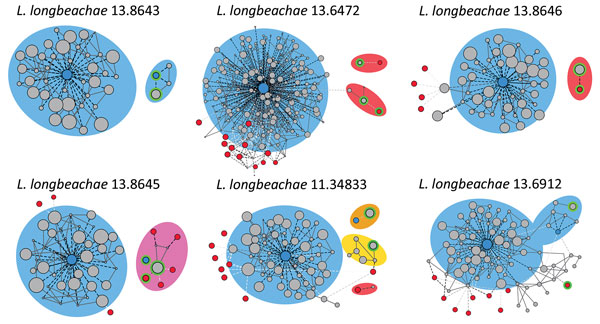
Figure 3. Legionella longbeachae plasmid analysis: contigs networks reconstructions for 6 representative L. longbeachae types of plasmid content. The networks of the contigs representing the main chromosome and plasmids comprising the genome obtained by using PLACNET (38), a program enabling reconstruction of plasmids from whole-genome sequence datasets. The sizes of the contig nodes (in gray) are proportional to their lengths; continuous lines correspond to scaffold links. Dashed lines represent BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) hits to the L. longbeachae (blue) or L. pneumophila (red) strains; intensity of the line is proportional to the hit (white indicates low, black indicates high). Green lines correspond to plasmid contigs. Background colors indicate species relatedness for the main chromosome and plasmids (blue for L. longbeachae, red for L. pneumophila, pink for a combination of both, and yellow for previously unidentified genomic content).
References
- Fields BS, Benson RF, Besser RE. Legionella and Legionnaires’ disease: 25 years of investigation. Clin Microbiol Rev. 2002;15:506–26. DOIPubMedGoogle Scholar
- European Centre for Disease Prevention and Control. Surveillance report. Legionnaires’ disease in Europe, 2010. 2012 [cited 2016 Jul 9]. http://ecdc.europa.eu/en/publications/publications/sur-legionnaires-disease-surveillance-2010.pdf
- Joseph CA, Ricketts KD; European Working Group for Legionella Infections. Legionnaires disease in Europe 2007-2008. Euro Surveill. 2010;15:19493.PubMedGoogle Scholar
- Marston BJ, Lipman HB, Breiman RF. Surveillance for Legionnaires’ disease. Risk factors for morbidity and mortality. Arch Intern Med. 1994;154:2417–22. DOIPubMedGoogle Scholar
- Li JS, O’Brien ED, Guest C. A review of national legionellosis surveillance in Australia, 1991 to 2000. Commun Dis Intell Q Rep. 2002;26:461–8.PubMedGoogle Scholar
- Cramp GJ, Harte D, Douglas NM, Graham F, Schousboe M, Sykes K. An outbreak of Pontiac fever due to Legionella longbeachae serogroup 2 found in potting mix in a horticultural nursery in New Zealand. Epidemiol Infect. 2010;138:15–20. DOIPubMedGoogle Scholar
- Whiley H, Bentham R. Legionella longbeachae and legionellosis. Emerg Infect Dis. 2011;17:579–83. DOIPubMedGoogle Scholar
- Yu VL, Plouffe JF, Pastoris MC, Stout JE, Schousboe M, Widmer A, et al. Distribution of Legionella species and serogroups isolated by culture in patients with sporadic community-acquired legionellosis: an international collaborative survey. J Infect Dis. 2002;186:127–8. DOIPubMedGoogle Scholar
- García C, Ugalde E, Campo AB, Miñambres E, Kovács N. Fatal case of community-acquired pneumonia caused by Legionella longbeachae in a patient with systemic lupus erythematosus. Eur J Clin Microbiol Infect Dis. 2004;23:116–8. DOIPubMedGoogle Scholar
- den Boer JW, Yzerman EPF, Jansen R, Bruin JP, Verhoef LPB, Neve G, et al. Legionnaires’ disease and gardening. Clin Microbiol Infect. 2007;13:88–91. DOIPubMedGoogle Scholar
- Potts A, Donaghy M, Marley M, Othieno R, Stevenson J, Hyland J, et al. Cluster of Legionnaires disease cases caused by Legionella longbeachae serogroup 1, Scotland, August to September 2013. Euro Surveill. 2013;18:20656. DOIPubMedGoogle Scholar
- Lindsay DSJ, Brown AW, Brown DJ, Pravinkumar SJ, Anderson E, Edwards GF. Legionella longbeachae serogroup 1 infections linked to potting compost. J Med Microbiol. 2012;61:218–22. DOIPubMedGoogle Scholar
- Steele TW, Lanser J, Sangster N. Isolation of Legionella longbeachae serogroup 1 from potting mixes. Appl Environ Microbiol. 1990;56:49–53.PubMedGoogle Scholar
- Koide M, Arakaki N, Saito A. Distribution of Legionella longbeachae and other legionellae in Japanese potting soils. J Infect Chemother. 2001;7:224–7. DOIPubMedGoogle Scholar
- Reuter S, Harrison TG, Köser CU, Ellington MJ, Smith GP, Parkhill J, et al. A pilot study of rapid whole-genome sequencing for the investigation of a Legionella outbreak. BMJ Open. 2013;3:e002175. DOIPubMedGoogle Scholar
- Rao C, Benhabib H, Ensminger AW. Phylogenetic reconstruction of the Legionella pneumophila Philadelphia-1 laboratory strains through comparative genomics. PLoS One. 2013;8:e64129. DOIPubMedGoogle Scholar
- Cazalet C, Gomez-Valero L, Rusniok C, Lomma M, Dervins-Ravault D, Newton HJ, et al. Analysis of the Legionella longbeachae genome and transcriptome uncovers unique strategies to cause Legionnaires’ disease. PLoS Genet. 2010;6:e1000851. DOIPubMedGoogle Scholar
- Kozak NA, Buss M, Lucas CE, Frace M, Govil D, Travis T, et al. Virulence factors encoded by Legionella longbeachae identified on the basis of the genome sequence analysis of clinical isolate D-4968. J Bacteriol. 2010;192:1030–44. DOIPubMedGoogle Scholar
- Gomez-Valero L, Rusniok C, Jarraud S, Vacherie B, Rouy Z, Barbe V, et al. Extensive recombination events and horizontal gene transfer shaped the Legionella pneumophila genomes. BMC Genomics. 2011;12:536. DOIPubMedGoogle Scholar
- Ratcliff RM, Lanser JA, Manning PA, Heuzenroeder MW. Sequence-based classification scheme for the genus Legionella targeting the mip gene. J Clin Microbiol. 1998;36:1560–7.PubMedGoogle Scholar
- Fallon RJ, Abraham WH. Experience with heat-killed antigens of L. longbeachae serogroups 1 and 2, and L. jordanis in the indirect fluorescence antibody test. Zentralbl Bakteriol Mikrobiol Hyg A. 1983;255:8–14.PubMedGoogle Scholar
- Babraham Bioinformatics. FastQC. 2010 [cited 2016 Jul 9]. http://www.bioinformatics.babraham.ac.uk/projects/fastqc
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–12.
- GitHub, Inc. wgsim Read Simulator [cited 2016 Jul 9]. https://github.com/lh3/wgsim
- Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42(D1):D633–42. DOIPubMedGoogle Scholar
- Kim M, Oh HS, Park SC, Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64:346–51. DOIPubMedGoogle Scholar
- Health Protection Scotland. Surveillance report: legionellosis in Scotland 2013–2014. 2015 Sep 1 [cited 2015 Aug 15]. http://www.hps.scot.nhs.uk/resp/wrdetail.aspx?id=65135&wrtype=6
- van der Mee-Marquet N, Domelier AS, Arnault L, Bloc D, Laudat P, Hartemann P, et al. Legionella anisa, a possible indicator of water contamination by Legionella pneumophila. J Clin Microbiol. 2006;44:56–9. DOIPubMedGoogle Scholar
- Svarrer CW, Uldum SA. The occurrence of Legionella species other than Legionella pneumophila in clinical and environmental samples in Denmark identified by mip gene sequencing and matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Microbiol Infect. 2012;18:1004–9. DOIPubMedGoogle Scholar
- Underwood AP, Jones G, Mentasti M, Fry NK, Harrison TG. Comparison of the Legionella pneumophila population structure as determined by sequence-based typing and whole genome sequencing. BMC Microbiol. 2013;13:302. DOIPubMedGoogle Scholar
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–67. DOIPubMedGoogle Scholar
- Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172:2665–81. DOIPubMedGoogle Scholar
- Marttinen P, Hanage WP, Croucher NJ, Connor TR, Harris SR, Bentley SD, et al. Detection of recombination events in bacterial genomes from large population samples. Nucleic Acids Res. 2012;40:e6. DOIPubMedGoogle Scholar
- de Been M, van Schaik W, Cheng L, Corander J, Willems RJ. Recent recombination events in the core genome are associated with adaptive evolution in Enterococcus faecium. Genome Biol Evol. 2013;5:1524–35. DOIPubMedGoogle Scholar
- Didelot X, Wilson DJ. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLOS Comput Biol. 2015;11:e1004041. DOIPubMedGoogle Scholar
- Coscollá M, Comas I, González-Candelas F. Quantifying nonvertical inheritance in the evolution of Legionella pneumophila. Mol Biol Evol. 2011;28:985–1001. DOIPubMedGoogle Scholar
- Sánchez-Busó L, Comas I, Jorques G, González-Candelas F. Recombination drives genome evolution in outbreak-related Legionella pneumophila isolates. Nat Genet. 2014;46:1205–11. DOIPubMedGoogle Scholar
- Lanza VF, de Toro M, Garcillán-Barcia MP, Mora A, Blanco J, Coque TM, et al. Plasmid flux in Escherichia coli ST131 sublineages, analyzed by plasmid constellation network (PLACNET), a new method for plasmid reconstruction from whole genome sequences. PLoS Genet. 2014;10:e1004766. DOIPubMedGoogle Scholar
- Cazalet C, Rusniok C, Brüggemann H, Zidane N, Magnier A, Ma L, et al. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat Genet. 2004;36:1165–73. DOIPubMedGoogle Scholar