Volume 23, Number 6—June 2017
Research Letter
Tick-Borne Encephalitis Virus in Ticks and Roe Deer, the Netherlands
Figure
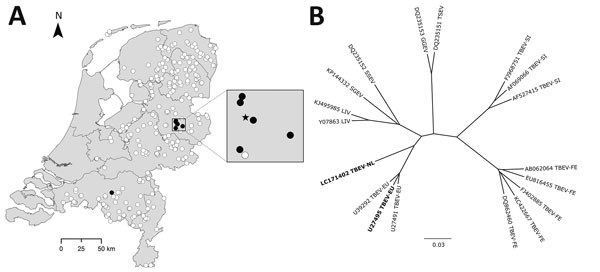
Figure. Spatial distribution of TBEV-positive roe deer and genetic cluster analysis of TBEV sequences from the Netherlands. A) Spatial distribution of serologic test results (solid black circle, SNT positive; open white circle, ELISA and/or SNT negative) for 297 serum samples from roe deer collected according to a sampling scheme designed to obtain a representative sample of the roe deer population from locations across the Netherlands. Enlargement of the National Park Sallandse Heuvelrug area indicates the locations of the TBEV serologically positive roe deer (solid black circle) in relation to the site with reverse transcription quantitative PCR–positive ticks (solid black star) from 2015. B) Genetic cluster analysis of TBEV-NL sequences obtained from tick pools in the Netherlands with other tickborne viruses (indicated by GenBank accession number). Bold indicates the TBEV-NL sequence, which consists of 10,242 nt of the genome (GenBank accession no. LC171402), and the TBEV-EU strain with which TBEV-NL clusters. Where available, representatives of the subtypes are included. We conducted distance-based analyses using Kimura 2-parameters distance estimates and constructed the trees using the neighbor-joining algorithm, implemented in Bionumerics 7.1 (Applied Math, Sint-Martens-Latem, Belgium). We calculated bootstrap proportions by analyzing 1,000 replicates for neighbor-joining trees. Scale bar indicates nucleotide substitutions per site. GGEV, Greek goat encephalomyelitis virus; LIV, Louping ill virus; SGEV, Spanish goat encephalitis virus; SSEV, Spanish sheep encephalitis virus; TBEV, tickborne encephalitis virus; TBEV-EU, TBEV European subtype; TBEV-FE, Far Eastern subtype; TBEV-NL, TBEV Netherlands subtype; TBEV-SI, TBEV Siberian subtype; TSEV, Turkish sheep encephalomyelitis virus.