Volume 23, Number 8—August 2017
Research
Genomic Characterization of Recrudescent Plasmodium malariae after Treatment with Artemether/Lumefantrine
Figure 3
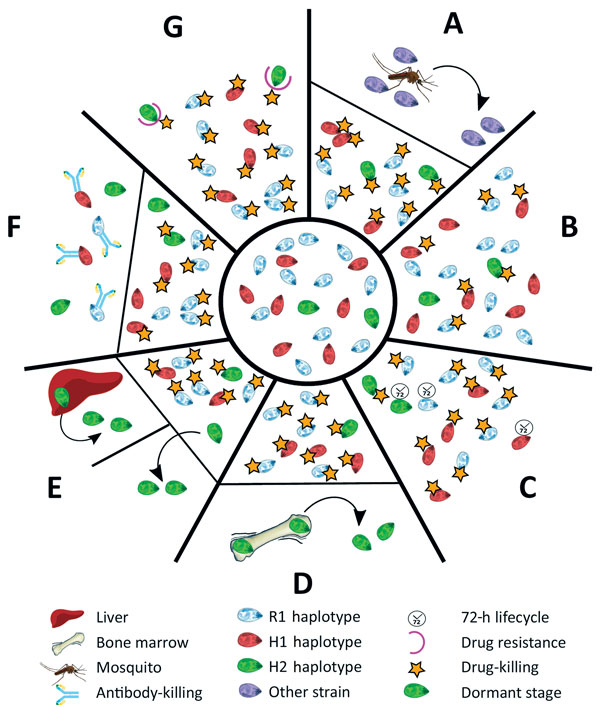
Figure 3. The different scenarios under which a second Plasmodium malariae infection could have occurred from the initial infection diagnosed in a patient at Royal Darwin Hospital, Darwin, Northern Territory, Australia, March–April 2015. Initial infection is shown in the inner circle. A) A completely new infection might have caused the second malaria onset. B) The drug might not have been absorbed at sufficient levels to kill all the parasites in the blood (pharmacokinetic cause). C) The longer intraerythrocytic life cycle of P. malariae (72 hours) might have enabled some parasites to survive the drug action until lumefantrine concentrations became subtherapeutic (pharmacokinetic cause). D) H2 parasites might have differentially sequestered with a biomass out of proportion with the peripheral parasitemia. E) Some parasites might have formed dormant stages in the liver, blood, or elsewhere (pharmacodynamic cause). F) An immune response might have been differentially primed against haplotypes at higher biomass. G) A haplotype within the initial infection might have been relatively drug resistant (fitness advantage). H1, haplotype 1; H2, haplotype 2; R1, reference genome.
1These authors contributed equally to this article.