Volume 24, Number 10—October 2018
CME ACTIVITY - Synopsis
Human Pegivirus in Patients with Encephalitis of Unclear Etiology, Poland
Figure 2
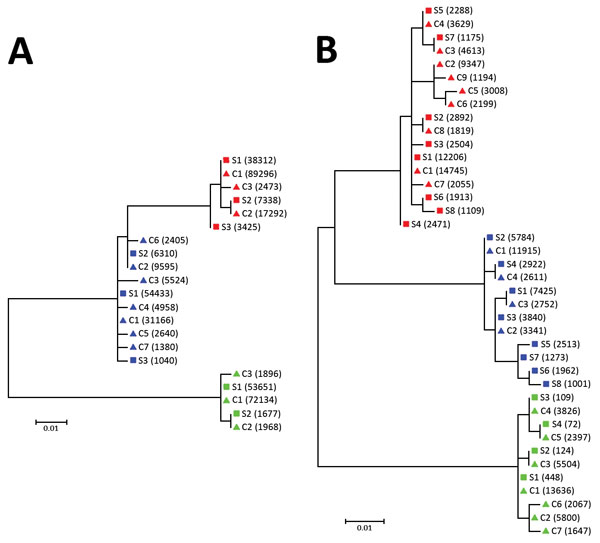
Figure 2. Phylogenetic analysis of A) 5′ UTR and B) E2 region sequences of human pegivirus from 3 patients with encephalitis of unclear origin, Poland, 2012–2015. Phylogenic trees were generated using ClustalX version 2.0 (http://www.clustal.org/clustal2/). Viral variant frequencies follow haplotype number. Red indicates patient 1; blue, patient 2; green, patient 3. Scale bars indicate number of nucleotide substitutions per site. C, cerebrospinal fluid; S, serum; UTR, untranslated region.
Page created: September 14, 2018
Page updated: September 14, 2018
Page reviewed: September 14, 2018
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.