Volume 24, Number 10—October 2018
Research
Frequent Genetic Mismatch between Vaccine Strains and Circulating Seasonal Influenza Viruses, Hong Kong, China, 1996–2012
Figure 2
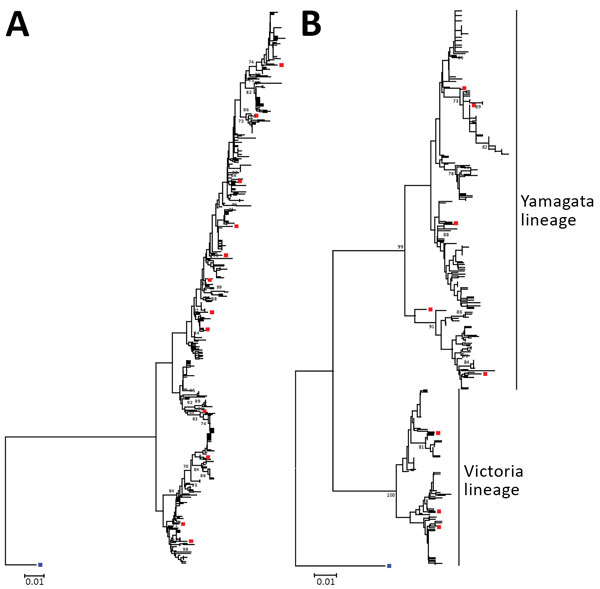
Figure 2. Neighbor-joining phylogenetic inference of near-complete hemagglutinin protein of influenza A(H3N2) (A) and influenza B (B), Hong Kong, China, 1996–2012. Pairwise protein sequence distances were calculated using a Poisson model. Blue squares denote ancestral influenza strains: influenza A(H3N2), A/Hong Kong/1/1968, GenBank accession no. CY044261; influenza B, B/Lee/1940, accession no. CY115111. Red squares denote recommended vaccine strains; for clarity, only those used in Figures 3 and 4 are shown. Bootstrap values >70% are shown at the respective nodes. All alignment positions containing gaps were omitted from the analysis. Trees were rooted to ancestral strains and drawn to scale. Scale bars indicate numbers of amino acid substitutions per position. GenBank accession numbers of hemagglutinin sequences of circulating strains collected in this study are shown in the Technical Appendix Table.
1Current affiliation: University of Alberta, Edmonton, Alberta, Canada.