Volume 24, Number 2—February 2018
Research
Lethal Respiratory Disease Associated with Human Rhinovirus C in Wild Chimpanzees, Uganda, 2013
Figure 2
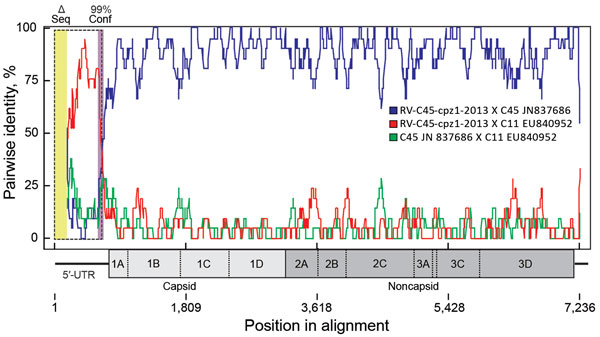
Figure 2. Recombination between viral genotypes rhinovirus C45 and C11 leading to RV-C45-cpz1-2013, the strain identified in the Kanyawara chimpanzee community, Uganda, 2013. Analyses were performed in RDP4 (17) on aligned rhinovirus C genome sequences of 36 known genotypes. Each alignment entry encoded the full or nearly full polyprotein gene sequence, but some sequences were missing fragments (<400 bp) of their respective 5′-UTRs (Δ seq, yellow box at left). The 3′ poly(A) tail was not included. A recombination event between the 2 viruses shown (GenBank nos. JN837686 and EU840952) is the most likely event among all full alignment comparisons (window size 20 bps) according to 6 of the 9 RDP4 algorithms. The average p values were RDP 2.8 × 10−81, GENECONV 3.0 × 10−70, MaxChi 1.4 × 10−18, Chimaera 2.1 × 10−21, SiScan 3.6 × 10–34, and 3Seq 1.5 × 10−27. BootScan, PhylPro, and LARD made no call for these particular parents. Purple box in the 5′-UTR denotes the 99% breakpoint confidence level (combined). Dashed box indicates the position of the most likely swapped fragment. The (Monte Carlo corrected) probability for this event is 2.8 × 10−81. The virus map is scaled to the alignment. Conf, confidence level; RV-C, rhinovirus C; Δ seq, missing sequence; UTR, untranslated region.
References
- Jacobs SE, Lamson DM, St George K, Walsh TJ. Human rhinoviruses. Clin Microbiol Rev. 2013;26:135–62. DOIPubMedGoogle Scholar
- Steinke JW, Borish L. Immune responses in rhinovirus-induced asthma exacerbations. Curr Allergy Asthma Rep. 2016;16:78. DOIPubMedGoogle Scholar
- Palmenberg AC, Gern JE. Classification and evolution of human rhinoviruses. Methods Mol Biol. 2015;1221:1–10. DOIPubMedGoogle Scholar
- Saraya T, Kurai D, Ishii H, Ito A, Sasaki Y, Niwa S, et al. Epidemiology of virus-induced asthma exacerbations: with special reference to the role of human rhinovirus. Front Microbiol. 2014;5:226. DOIPubMedGoogle Scholar
- Bizzintino J, Lee WM, Laing IA, Vang F, Pappas T, Zhang G, et al. Association between human rhinovirus C and severity of acute asthma in children. Eur Respir J. 2011;37:1037–42. DOIPubMedGoogle Scholar
- Liu Y, Hill MG, Klose T, Chen Z, Watters K, Bochkov YA, et al. Atomic structure of a rhinovirus C, a virus species linked to severe childhood asthma. Proc Natl Acad Sci U S A. 2016;113:8997–9002. DOIPubMedGoogle Scholar
- Bochkov YA, Watters K, Ashraf S, Griggs TF, Devries MK, Jackson DJ, et al. Cadherin-related family member 3, a childhood asthma susceptibility gene product, mediates rhinovirus C binding and replication. Proc Natl Acad Sci U S A. 2015;112:5485–90. DOIPubMedGoogle Scholar
- Mathieson I, Lazaridis I, Rohland N, Mallick S, Patterson N, Roodenberg SA, et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature. 2015;528:499–503. DOIPubMedGoogle Scholar
- Palmenberg AC. Rhinovirus C, asthma, and cell surface expression of virus receptor CDHR3. J Virol. 2017;91:e00072-17. DOIPubMedGoogle Scholar
- Sharp PM, Rayner JC, Hahn BH. Evolution. Great apes and zoonoses. Science. 2013;340:284–6. DOIPubMedGoogle Scholar
- Köndgen S, Kühl H, N’Goran PK, Walsh PD, Schenk S, Ernst N, et al. Pandemic human viruses cause decline of endangered great apes. Curr Biol. 2008;18:260–4. DOIPubMedGoogle Scholar
- Köndgen S, Schenk S, Pauli G, Boesch C, Leendertz FH. Noninvasive monitoring of respiratory viruses in wild chimpanzees. EcoHealth. 2010;7:332–41. DOIPubMedGoogle Scholar
- Palacios G, Lowenstine LJ, Cranfield MR, Gilardi KV, Spelman L, Lukasik-Braum M, et al. Human metapneumovirus infection in wild mountain gorillas, Rwanda. Emerg Infect Dis. 2011;17:711–3. DOIPubMedGoogle Scholar
- Grützmacher KS, Köndgen S, Keil V, Todd A, Feistner A, Herbinger I, et al. Codetection of respiratory syncytial virus in habituated wild western lowland gorillas and humans during a respiratory disease outbreak. EcoHealth. 2016;13:499–510. DOIPubMedGoogle Scholar
- Toohey-Kurth K, Sibley SD, Goldberg TL. Metagenomic assessment of adventitious viruses in commercial bovine sera. Biologicals. 2017;47:64–8. DOIPubMedGoogle Scholar
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4. DOIPubMedGoogle Scholar
- Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015;1:vev003. DOIPubMedGoogle Scholar
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–66. DOIPubMedGoogle Scholar
- Abascal F, Zardoya R, Telford MJ. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010;38(suppl_2):W7-13. DOIPubMedGoogle Scholar
- Talavera G, Castresana J, Kjer K, Page R, Sullivan J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–77. DOIPubMedGoogle Scholar
- Santorum JM, Darriba D, Taboada GL, Posada D. jmodeltest.org: selection of nucleotide substitution models on the cloud. Bioinformatics. 2014;30:1310–1. DOIPubMedGoogle Scholar
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21. DOIPubMedGoogle Scholar
- Krunic N, Merante F, Yaghoubian S, Himsworth D, Janeczko R. Advances in the diagnosis of respiratory tract infections: role of the Luminex xTAG respiratory viral panel. Ann N Y Acad Sci. 2011;1222:6–13. DOIPubMedGoogle Scholar
- Bochkov YA, Gern JE. Clinical and molecular features of human rhinovirus C. Microbes Infect. 2012;14:485–94. DOIPubMedGoogle Scholar
- Prado-Martinez J, Sudmant PH, Kidd JM, Li H, Kelley JL, Lorente-Galdos B, et al. Great ape genetic diversity and population history. Nature. 2013;499:471–5. DOIPubMedGoogle Scholar
- Althaus CL. Estimating the reproduction number of Ebola virus (EBOV) during the 2014 outbreak in West Africa. PLoS Curr. 2014;6:6.PubMedGoogle Scholar
- Sikazwe CT, Chidlow GR, Imrie A, Smith DW. Reliable quantification of rhinovirus species C using real-time PCR. J Virol Methods. 2016;235:65–72. DOIPubMedGoogle Scholar
- Bochkov YA, Grindle K, Vang F, Evans MD, Gern JE. Improved molecular typing assay for rhinovirus species A, B, and C. J Clin Microbiol. 2014;52:2461–71. DOIPubMedGoogle Scholar
- Harvala H, McIntyre CL, Imai N, Clasper L, Djoko CF, LeBreton M, et al. High seroprevalence of enterovirus infections in apes and old world monkeys. Emerg Infect Dis. 2012;18:283–6. DOIPubMedGoogle Scholar
- Sadeuh-Mba SA, Bessaud M, Joffret ML, Endegue Zanga MC, Balanant J, Mpoudi Ngole E, et al. Characterization of Enteroviruses from non-human primates in cameroon revealed virus types widespread in humans along with candidate new types and species. PLoS Negl Trop Dis. 2014;8:e3052. DOIPubMedGoogle Scholar
- Lu X, Erdman DD. Molecular typing of human adenoviruses by PCR and sequencing of a partial region of the hexon gene. Arch Virol. 2006;151:1587–602. DOIPubMedGoogle Scholar
- Nkogue CN, Horie M, Fujita S, Ogino M, Kobayashi Y, Mizukami K, et al. Molecular epidemiological study of adenovirus infecting western lowland gorillas and humans in and around Moukalaba-Doudou National Park (Gabon). Virus Genes. 2016;52:671–8. DOIPubMedGoogle Scholar
- Seimon TA, Olson SH, Lee KJ, Rosen G, Ondzie A, Cameron K, et al. Adenovirus and herpesvirus diversity in free-ranging great apes in the Sangha region of the Republic Of Congo. PLoS One. 2015;10:e0118543. DOIPubMedGoogle Scholar
- Wevers D, Metzger S, Babweteera F, Bieberbach M, Boesch C, Cameron K, et al. Novel adenoviruses in wild primates: a high level of genetic diversity and evidence of zoonotic transmissions. J Virol. 2011;85:10774–84. DOIPubMedGoogle Scholar
- Hoppe E, Pauly M, Gillespie TR, Akoua-Koffi C, Hohmann G, Fruth B, et al. Multiple cross-species transmission events of human adenoviruses (HAdV) during hominine evolution. Mol Biol Evol. 2015;32:2072–84. DOIPubMedGoogle Scholar
- Dick EC. Experimental infections of chimpanzees with human rhinovirus types 14 and 43. Proc Soc Exp Biol Med. 1968;127:1079–81. DOIPubMedGoogle Scholar
- Dick EC, Dick CR. A subclinical outbreak of human rhinovirus 31 infection in chimpanzees. Am J Epidemiol. 1968;88:267–72. DOIPubMedGoogle Scholar
- Goodall J. The chimpanzees of Gombe: patterns of behavior. Cambridge: Harvard University Press; 1986.
- Eggo RM, Scott JG, Galvani AP, Meyers LA. Respiratory virus transmission dynamics determine timing of asthma exacerbation peaks: Evidence from a population-level model. Proc Natl Acad Sci U S A. 2016;113:2194–9. DOIPubMedGoogle Scholar
- Williams JM, Lonsdorf EV, Wilson ML, Schumacher-Stankey J, Goodall J, Pusey AE. Causes of death in the Kasekela chimpanzees of Gombe National Park, Tanzania. Am J Primatol. 2008;70:766–77. DOIPubMedGoogle Scholar
- Griggs TF, Bochkov YA, Nakagome K, Palmenberg AC, Gern JE. Production, purification, and capsid stability of rhinovirus C types. J Virol Methods. 2015;217:18–23. DOIPubMedGoogle Scholar
- Honkanen H, Oikarinen S, Peltonen P, Simell O, Ilonen J, Veijola R, et al. Human rhinoviruses including group C are common in stool samples of young Finnish children. J Clin Virol. 2013;56:250–4. DOIPubMedGoogle Scholar
- Roy S, Vandenberghe LH, Kryazhimskiy S, Grant R, Calcedo R, Yuan X, et al. Isolation and characterization of adenoviruses persistently shed from the gastrointestinal tract of non-human primates. PLoS Pathog. 2009;5:e1000503. DOIPubMedGoogle Scholar
- Keele BF, Jones JH, Terio KA, Estes JD, Rudicell RS, Wilson ML, et al. Increased mortality and AIDS-like immunopathology in wild chimpanzees infected with SIVcpz. Nature. 2009;460:515–9. DOIPubMedGoogle Scholar
- Santiago ML, Lukasik M, Kamenya S, Li Y, Bibollet-Ruche F, Bailes E, et al. Foci of endemic simian immunodeficiency virus infection in wild-living eastern chimpanzees (Pan troglodytes schweinfurthii). J Virol. 2003;77:7545–62. DOIPubMedGoogle Scholar
- Gilardi KV, Gillespie TR, Leendertz FH, Macfie EJ, Travis DA, Whittier CA, et al. Best practice guidelines for health monitoring and disease control in great ape populations. Gland (Switzerland): IUCN Special Survival Commission Primate Specialist Group; 2015.
- Reagan KJ, McGeady ML, Crowell RL. Persistence of human rhinovirus infectivity under diverse environmental conditions. Appl Environ Microbiol. 1981;41:618–20.PubMedGoogle Scholar
- Bischoff WE, Tucker BK, Wallis ML, Reboussin BA, Pfaller MA, Hayden FG, et al. Preventing the airborne spread of Staphylococcus aureus by persons with the common cold: effect of surgical scrubs, gowns, and masks. Infect Control Hosp Epidemiol. 2007;28:1148–54. DOIPubMedGoogle Scholar
- Turner RB, Fuls JL, Rodgers ND. Effectiveness of hand sanitizers with and without organic acids for removal of rhinovirus from hands. Antimicrob Agents Chemother. 2010;54:1363–4. DOIPubMedGoogle Scholar
- Huguenel ED, Cohn D, Dockum DP, Greve JM, Fournel MA, Hammond L, et al. Prevention of rhinovirus infection in chimpanzees by soluble intercellular adhesion molecule-1. Am J Respir Crit Care Med. 1997;155:1206–10. DOIPubMedGoogle Scholar