Volume 24, Number 2—February 2018
Research
Lethal Respiratory Disease Associated with Human Rhinovirus C in Wild Chimpanzees, Uganda, 2013
Figure 3
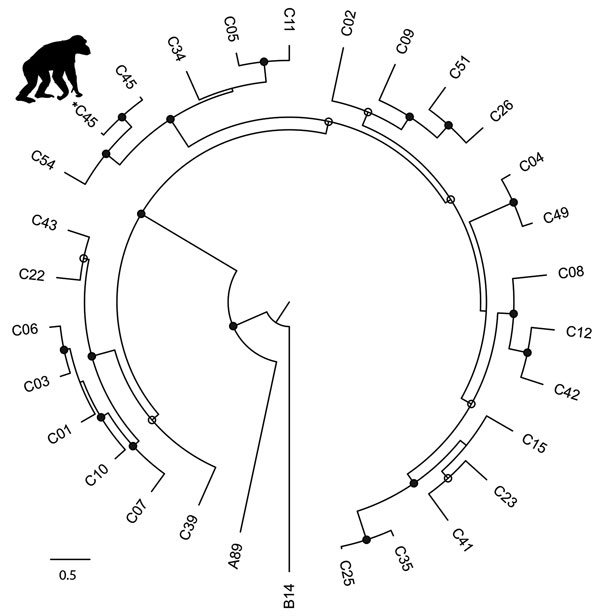
Figure 3. Phylogenetic tree of rhinovirus C variants. The tree was constructed from a codon-based alignment (6,234 positions) of the new chimpanzee-derived sequence identified in the Kanyawara chimpanzee community, Uganda, 2013 (indicated by the asterisk and chimpanzee silhouette), and all human-derived RV-C complete polyprotein gene sequences available in GenBank as of December 18, 2016, with rhinoviruses A and B from the RefSeq database included as outgroups. We created alignments using the MAFFT algorithm (18) implemented in the computer program Translator X (19), with the Gblocks algorithm (20) applied to remove poorly aligned regions. We constructed trees using the maximum-likelihood method implemented in PhyML (22), with best-fit models of molecular evolution estimated from the data by using jModelTest (21). Circles on nodes indicate statistical confidence on the basis of 1,000 bootstrap replicates of the data (closed circles 100%; open circles >75%). Scale bar indicates nucleotide substitutions per site. GenBank accession numbers and other details of the RV-C sequences included in the analysis are in Technical Appendix Table).
References
- Jacobs SE, Lamson DM, St George K, Walsh TJ. Human rhinoviruses. Clin Microbiol Rev. 2013;26:135–62. DOIPubMedGoogle Scholar
- Steinke JW, Borish L. Immune responses in rhinovirus-induced asthma exacerbations. Curr Allergy Asthma Rep. 2016;16:78. DOIPubMedGoogle Scholar
- Palmenberg AC, Gern JE. Classification and evolution of human rhinoviruses. Methods Mol Biol. 2015;1221:1–10. DOIPubMedGoogle Scholar
- Saraya T, Kurai D, Ishii H, Ito A, Sasaki Y, Niwa S, et al. Epidemiology of virus-induced asthma exacerbations: with special reference to the role of human rhinovirus. Front Microbiol. 2014;5:226. DOIPubMedGoogle Scholar
- Bizzintino J, Lee WM, Laing IA, Vang F, Pappas T, Zhang G, et al. Association between human rhinovirus C and severity of acute asthma in children. Eur Respir J. 2011;37:1037–42. DOIPubMedGoogle Scholar
- Liu Y, Hill MG, Klose T, Chen Z, Watters K, Bochkov YA, et al. Atomic structure of a rhinovirus C, a virus species linked to severe childhood asthma. Proc Natl Acad Sci U S A. 2016;113:8997–9002. DOIPubMedGoogle Scholar
- Bochkov YA, Watters K, Ashraf S, Griggs TF, Devries MK, Jackson DJ, et al. Cadherin-related family member 3, a childhood asthma susceptibility gene product, mediates rhinovirus C binding and replication. Proc Natl Acad Sci U S A. 2015;112:5485–90. DOIPubMedGoogle Scholar
- Mathieson I, Lazaridis I, Rohland N, Mallick S, Patterson N, Roodenberg SA, et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature. 2015;528:499–503. DOIPubMedGoogle Scholar
- Palmenberg AC. Rhinovirus C, asthma, and cell surface expression of virus receptor CDHR3. J Virol. 2017;91:e00072-17. DOIPubMedGoogle Scholar
- Sharp PM, Rayner JC, Hahn BH. Evolution. Great apes and zoonoses. Science. 2013;340:284–6. DOIPubMedGoogle Scholar
- Köndgen S, Kühl H, N’Goran PK, Walsh PD, Schenk S, Ernst N, et al. Pandemic human viruses cause decline of endangered great apes. Curr Biol. 2008;18:260–4. DOIPubMedGoogle Scholar
- Köndgen S, Schenk S, Pauli G, Boesch C, Leendertz FH. Noninvasive monitoring of respiratory viruses in wild chimpanzees. EcoHealth. 2010;7:332–41. DOIPubMedGoogle Scholar
- Palacios G, Lowenstine LJ, Cranfield MR, Gilardi KV, Spelman L, Lukasik-Braum M, et al. Human metapneumovirus infection in wild mountain gorillas, Rwanda. Emerg Infect Dis. 2011;17:711–3. DOIPubMedGoogle Scholar
- Grützmacher KS, Köndgen S, Keil V, Todd A, Feistner A, Herbinger I, et al. Codetection of respiratory syncytial virus in habituated wild western lowland gorillas and humans during a respiratory disease outbreak. EcoHealth. 2016;13:499–510. DOIPubMedGoogle Scholar
- Toohey-Kurth K, Sibley SD, Goldberg TL. Metagenomic assessment of adventitious viruses in commercial bovine sera. Biologicals. 2017;47:64–8. DOIPubMedGoogle Scholar
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4. DOIPubMedGoogle Scholar
- Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015;1:vev003. DOIPubMedGoogle Scholar
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–66. DOIPubMedGoogle Scholar
- Abascal F, Zardoya R, Telford MJ. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010;38(suppl_2):W7-13. DOIPubMedGoogle Scholar
- Talavera G, Castresana J, Kjer K, Page R, Sullivan J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–77. DOIPubMedGoogle Scholar
- Santorum JM, Darriba D, Taboada GL, Posada D. jmodeltest.org: selection of nucleotide substitution models on the cloud. Bioinformatics. 2014;30:1310–1. DOIPubMedGoogle Scholar
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21. DOIPubMedGoogle Scholar
- Krunic N, Merante F, Yaghoubian S, Himsworth D, Janeczko R. Advances in the diagnosis of respiratory tract infections: role of the Luminex xTAG respiratory viral panel. Ann N Y Acad Sci. 2011;1222:6–13. DOIPubMedGoogle Scholar
- Bochkov YA, Gern JE. Clinical and molecular features of human rhinovirus C. Microbes Infect. 2012;14:485–94. DOIPubMedGoogle Scholar
- Prado-Martinez J, Sudmant PH, Kidd JM, Li H, Kelley JL, Lorente-Galdos B, et al. Great ape genetic diversity and population history. Nature. 2013;499:471–5. DOIPubMedGoogle Scholar
- Althaus CL. Estimating the reproduction number of Ebola virus (EBOV) during the 2014 outbreak in West Africa. PLoS Curr. 2014;6:6.PubMedGoogle Scholar
- Sikazwe CT, Chidlow GR, Imrie A, Smith DW. Reliable quantification of rhinovirus species C using real-time PCR. J Virol Methods. 2016;235:65–72. DOIPubMedGoogle Scholar
- Bochkov YA, Grindle K, Vang F, Evans MD, Gern JE. Improved molecular typing assay for rhinovirus species A, B, and C. J Clin Microbiol. 2014;52:2461–71. DOIPubMedGoogle Scholar
- Harvala H, McIntyre CL, Imai N, Clasper L, Djoko CF, LeBreton M, et al. High seroprevalence of enterovirus infections in apes and old world monkeys. Emerg Infect Dis. 2012;18:283–6. DOIPubMedGoogle Scholar
- Sadeuh-Mba SA, Bessaud M, Joffret ML, Endegue Zanga MC, Balanant J, Mpoudi Ngole E, et al. Characterization of Enteroviruses from non-human primates in cameroon revealed virus types widespread in humans along with candidate new types and species. PLoS Negl Trop Dis. 2014;8:e3052. DOIPubMedGoogle Scholar
- Lu X, Erdman DD. Molecular typing of human adenoviruses by PCR and sequencing of a partial region of the hexon gene. Arch Virol. 2006;151:1587–602. DOIPubMedGoogle Scholar
- Nkogue CN, Horie M, Fujita S, Ogino M, Kobayashi Y, Mizukami K, et al. Molecular epidemiological study of adenovirus infecting western lowland gorillas and humans in and around Moukalaba-Doudou National Park (Gabon). Virus Genes. 2016;52:671–8. DOIPubMedGoogle Scholar
- Seimon TA, Olson SH, Lee KJ, Rosen G, Ondzie A, Cameron K, et al. Adenovirus and herpesvirus diversity in free-ranging great apes in the Sangha region of the Republic Of Congo. PLoS One. 2015;10:e0118543. DOIPubMedGoogle Scholar
- Wevers D, Metzger S, Babweteera F, Bieberbach M, Boesch C, Cameron K, et al. Novel adenoviruses in wild primates: a high level of genetic diversity and evidence of zoonotic transmissions. J Virol. 2011;85:10774–84. DOIPubMedGoogle Scholar
- Hoppe E, Pauly M, Gillespie TR, Akoua-Koffi C, Hohmann G, Fruth B, et al. Multiple cross-species transmission events of human adenoviruses (HAdV) during hominine evolution. Mol Biol Evol. 2015;32:2072–84. DOIPubMedGoogle Scholar
- Dick EC. Experimental infections of chimpanzees with human rhinovirus types 14 and 43. Proc Soc Exp Biol Med. 1968;127:1079–81. DOIPubMedGoogle Scholar
- Dick EC, Dick CR. A subclinical outbreak of human rhinovirus 31 infection in chimpanzees. Am J Epidemiol. 1968;88:267–72. DOIPubMedGoogle Scholar
- Goodall J. The chimpanzees of Gombe: patterns of behavior. Cambridge: Harvard University Press; 1986.
- Eggo RM, Scott JG, Galvani AP, Meyers LA. Respiratory virus transmission dynamics determine timing of asthma exacerbation peaks: Evidence from a population-level model. Proc Natl Acad Sci U S A. 2016;113:2194–9. DOIPubMedGoogle Scholar
- Williams JM, Lonsdorf EV, Wilson ML, Schumacher-Stankey J, Goodall J, Pusey AE. Causes of death in the Kasekela chimpanzees of Gombe National Park, Tanzania. Am J Primatol. 2008;70:766–77. DOIPubMedGoogle Scholar
- Griggs TF, Bochkov YA, Nakagome K, Palmenberg AC, Gern JE. Production, purification, and capsid stability of rhinovirus C types. J Virol Methods. 2015;217:18–23. DOIPubMedGoogle Scholar
- Honkanen H, Oikarinen S, Peltonen P, Simell O, Ilonen J, Veijola R, et al. Human rhinoviruses including group C are common in stool samples of young Finnish children. J Clin Virol. 2013;56:250–4. DOIPubMedGoogle Scholar
- Roy S, Vandenberghe LH, Kryazhimskiy S, Grant R, Calcedo R, Yuan X, et al. Isolation and characterization of adenoviruses persistently shed from the gastrointestinal tract of non-human primates. PLoS Pathog. 2009;5:e1000503. DOIPubMedGoogle Scholar
- Keele BF, Jones JH, Terio KA, Estes JD, Rudicell RS, Wilson ML, et al. Increased mortality and AIDS-like immunopathology in wild chimpanzees infected with SIVcpz. Nature. 2009;460:515–9. DOIPubMedGoogle Scholar
- Santiago ML, Lukasik M, Kamenya S, Li Y, Bibollet-Ruche F, Bailes E, et al. Foci of endemic simian immunodeficiency virus infection in wild-living eastern chimpanzees (Pan troglodytes schweinfurthii). J Virol. 2003;77:7545–62. DOIPubMedGoogle Scholar
- Gilardi KV, Gillespie TR, Leendertz FH, Macfie EJ, Travis DA, Whittier CA, et al. Best practice guidelines for health monitoring and disease control in great ape populations. Gland (Switzerland): IUCN Special Survival Commission Primate Specialist Group; 2015.
- Reagan KJ, McGeady ML, Crowell RL. Persistence of human rhinovirus infectivity under diverse environmental conditions. Appl Environ Microbiol. 1981;41:618–20.PubMedGoogle Scholar
- Bischoff WE, Tucker BK, Wallis ML, Reboussin BA, Pfaller MA, Hayden FG, et al. Preventing the airborne spread of Staphylococcus aureus by persons with the common cold: effect of surgical scrubs, gowns, and masks. Infect Control Hosp Epidemiol. 2007;28:1148–54. DOIPubMedGoogle Scholar
- Turner RB, Fuls JL, Rodgers ND. Effectiveness of hand sanitizers with and without organic acids for removal of rhinovirus from hands. Antimicrob Agents Chemother. 2010;54:1363–4. DOIPubMedGoogle Scholar
- Huguenel ED, Cohn D, Dockum DP, Greve JM, Fournel MA, Hammond L, et al. Prevention of rhinovirus infection in chimpanzees by soluble intercellular adhesion molecule-1. Am J Respir Crit Care Med. 1997;155:1206–10. DOIPubMedGoogle Scholar