Volume 24, Number 2—February 2018
Research
Plasmid-Encoded Transferable mecB-Mediated Methicillin Resistance in Staphylococcus aureus
Cite This Article
Citation for Media
Abstract
During cefoxitin-based nasal screening, phenotypically categorized methicillin-resistant Staphylococcus aureus (MRSA) was isolated and tested negative for the presence of the mecA and mecC genes as well as for the SCCmec-orfX junction region. The isolate was found to carry a mecB gene previously described for Macrococcus caseolyticus but not for staphylococcal species. The gene is flanked by β-lactam regulatory genes similar to mecR, mecI, and blaZ and is part of an 84.6-kb multidrug-resistance plasmid that harbors genes encoding additional resistances to aminoglycosides (aacA-aphD, aphA, and aadK) as well as macrolides (ermB) and tetracyclines (tetS). This further plasmidborne β-lactam resistance mechanism harbors the putative risk of acceleration or reacceleration of MRSA spread, resulting in broad ineffectiveness of β-lactams as a main therapeutic application against staphylococcal infections.
Staphylococcal cassette chromosome mec (SCCmec)–mediated β-lactam resistance resulting from production of an additional penicillin-binding protein (PBP) 2a drastically limits the treatment options in cases of hospital- and community-related infections by staphylococci, leading to increased illness, death, and socioeconomic costs (1,2). Besides methicillin-resistant coagulase-negative staphylococci, notorious for foreign body–associated infections, methicillin-resistant Staphylococcus aureus (MRSA) strains are a global public health priority, despite some countries in Europe reporting stabilizing or decreasing MRSA rates (3–5). Since the initial reports of MRSA in 1961, several epidemic waves have resulted in threats of healthcare-, community-, and livestock-associated MRSA (6–9).
For staphylococci, 2 PBP 2a-encoding genes, mecA and mecC, including several allotypes, have been described as chromosomally located genetic bases for phenotypic methicillin resistance (10–14). In contrast, mecB, originally described as mecAm, was reported as part of a probable primordial form of a methicillin resistance gene complex often found in a transposon mec complex (Tn6045) in Macrococcus caseolyticus, a colonizer of animal skin (15,16). Just recently, a mecD gene, most closely related to mecB, has been detected in bovine and canine M. caseolyticus isolates (17).
The impact of plasmidborne resistance for staphylococci is abundantly demonstrated for β-lactamase–mediated penicillin resistance. Resistance rates are >60% in human S. aureus isolates from the general population and >90% from hospital-related cases, regardless of the clinical background (18,19). In contrast to frequent interstrain and interspecies transmission of resistance plasmids by conjugation or transduction, only a relatively low rate of spontaneous horizontal transfer of SCCmec elements is assumed, resulting in still-manageable and controllable MRSA rates if prevention measures are adequate (20–24). However, transferable methicillin resistance might bear the consequence of an almost complete loss of β-lactam drugs as the most efficient class of antibacterial drugs for treatment of staphylococcal infections. Here, we report both a plasmid-encoded, and thereby transferable, methicillin resistance encoded by mecB and the occurrence of this gene in an isolate of the genus Staphylococcus.
Strain Detection and Identification
At the University Hospital of Münster, Germany, MRSA is generally cultured, identified, and differentiated by routine microbiological diagnostic methods using dextrose broth enrichment; chromID MRSA selective agar (bioMérieux, Marcy-l’Étoile, France), which contains cefoxitin; VITEK 2 automated system (bioMérieux) applying the antimicrobial susceptibility test card AST-P632; PBP2a detection kit (PBP2a Culture Colony Test, Alere, San Diego, CA, USA); S. aureus–specific PCR targeting mecA/mecC (GenoType MRSA, Hain-Lifescience, Nehren, Germany); and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (Microflex-LT system, MALDI-Biotyper 3.0; Bruker Daltonik, Bremen, Germany). In February 2016, an S. aureus isolate (which we numbered UKM4229) was recovered during routine MRSA screening. The isolate displayed a β-lactam–resistant phenotype without carrying the methicillin resistance genes mecA or mecC. For further characterizations, isolate UKM4229 was stored at −80°C and was cultivated on chromID MRSA agar (bioMérieux) at 37°C.
Genetic Analysis
We extracted genomic DNA from S. aureus isolate UKM4229 using the QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. We isolated plasmid DNA with the PrepEase MiniSpin Plasmid Kit (Affymetrix USB, Santa Clara, CA, USA) following the protocol standards. For both plasmid and genomic DNA, we applied lysostaphin (20 µg/mL) (Wakchemie, Steinbach, Germany) for bacterial cell lysis. We performed multilocus sequence typing and spa gene typing initially as described elsewhere (25,26) and confirmed our results later by analysis of whole-genome sequencing (WGS) and DNA microarray data (discussed later in this article). We analyzed DNA sequences using RidomStaphType and SeqSphere+ (Ridom GmbH, Münster, Germany). Applying DNA microarray analysis (IdentiBAC Microarray; Alere Technologies GmbH, Jena, Germany), we identified resistance and virulence determinants and checked genotyping results.
Molecular Confirmation of Methicillin Resistance
Using PCR, we tested for the presence of methicillin resistance genes mecA and mecC (27,28) as well as mecB. DNA sequences of PCR oligonucleotides are given in Table 1. Oligonucleotides for mecB were made on basis of the plasmid pMCCL2 of M. caseolyticus (GenBank accession no. NC_011996.1). We performed PCR reactions using the following protocol for mecA: 5 min at 95°C; 40 cycles of 0.5 min at 95°C, 0.5 min at 55.5°C, and 0.75 min at 72°C; and final elongation of 7 min at 72°C. The protocol for mecB was 5 min at 95°C; 35 cycles of 0.5 min at 95°C, 0.5 min at 57°C, and 2.5 min at 72°C; and final elongation of 7 min at 72°C. The protocol for mecC: 5 min at 95°C; 40 cycles of 0.5 min at 95°C, 0.5 min at 59.3°C, and 2 min at 72°C; and final elongation of 7 min at 72°C.
Antibiotic Drug Susceptibility Testing
We determined the MIC of cefoxitin for S. aureus isolate UKM4229 by the reference broth microdilution method according to the International Organization for Standardization (ISO) 20776-1 guideline (https://www.iso.org/standard/41630.html), as required by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) and the Clinical and Laboratory Standards Institute (CLSI). Cefoxitin (Sigma Aldrich, Taufkirchen, Germany) was tested in 2-fold concentrations (0.25–128 µg/mL). We subcultured the isolate and incubated it overnight before testing.
We investigated the susceptibility profile of UKM4229 by determining MICs of various β-lactam and non–β-lactam antibiotic drugs (Table 2) using the gradient diffusion method (Etest; bioMérieux) according to the manufacturer’s instructions. As recommended, the inoculated plates were incubated at 35°C for 18 ± 2 hours. In addition, we tested oxacillin using conditions for increased expression of methicillin resistance, as reported for mecA isolates (30): Mueller-Hinton agar supplemented with 2% saline, incubation at 30°C, and prolonged incubation up to 48 h. We investigated the applicability of a commercial automated susceptibility testing device to recognize methicillin resistance due to presence of mecB in S. aureus by using the VITEK 2 system. We evaluated the in vitro activity of the endolysin HY-133 against UKM4229 using the broth microdilution method in accordance with ISO 20776-1 guidance (https://www.iso.org/standard/41630.html), as described elsewhere (31,32). In brief, we tested 2-fold final concentrations of HY-133 ranging from 0.06 µg/mL to 8 µg/mL using 1–5 × 105 CFU/mL suspension of UKM4229 in cation-adjusted Mueller-Hinton broth. The MICs were read after incubation at 35°C for 18 ± 2 h.
We performed all experiments in triplicate on different days and calculated the median MIC values. We used S. aureus ATCC 29213 as a quality control strain on every testing day. For the antibiotic drugs we used, the MICs for the quality control strain were within acceptable limits throughout the testing.
Whole-Genome Sequencing
For the PacBio RS II platform (Pacific Biosciences, Menlo Park, CA, USA), we extracted staphylococcal DNA using the Genomic-tip 20/G Kit (QIAGEN) according to the manufacturer’s instructions, excep that we applied lysostaphin (20 µg/mL) (Wakchemie) for bacterial cell lysis. We sequenced the extracted high-quality, double-stranded DNA (5 µg) using P6-C4 chemistry on the PacBio RS II instrumentation using 4-hour movie collection and 110 pmol/L of complexed 20-kb SMRTbell library. We performed the initial de novo assembly using the HGAP3 v2.3.0 Assembler (Icahn Institute for Genomics and Multiscale Biology, Icahn School of Medicine at Mount Sinai, New York, NY, USA). We annotated the assembled genome through the GenDB pipeline (33). We verified questionable sequences within the plasmids by applying PCR (LA-Taq-DNA-Polymerase; Takara, Frankfurt am Main, Germany) and Sanger sequencing (Eurofins Genomics, Ebersberg, Germany).
During routine MRSA screening, we recovered an S. aureus isolate UKM4229 from a combined nasal-throat swab of a 67-year-old male cardiology inpatient who had no signs of infection. We isolated colonies with typical appearance for presumptive MRSA from a chromogenic MRSA selective agar and identified them as S. aureus by VITEK 2, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry, and PCR. Discrepancies between phenotypic detection of methicillin resistance by VITEK 2 and negative results of a PBP2a detection kit, as well as negative mecA and mecC test results, by commercial and in-house PCRs led to the detection of a mecB-encoded methicillin resistance.
S. aureus isolate UKM4229 showed a median MIC of 32 µg/mL for cefoxitin, as determined by broth microdilution and gradient diffusion tests. The MICs of other antibiotics, as well as correspondent interpretative categories, are shown in Table 2; the resistance gene profile is given in Technical Appendix Table 1. Optimal oxacillin testing conditions previously reported to increase expression of methicillin resistance in mecA isolates (30) unexpectedly led to lower oxacillin MIC values for UKM4229 (Table 2). A novel anti–S. aureus agent in development, the recombinant phage endolysin HY-133 (Hyglos, Bernried, Germany) (31,32), was also active. VITEK 2 recognized mecB-associated methicillin resistance by oxacillin MIC determination and cefoxitin screening.
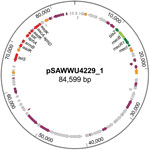
Although mecA/mecC PCR did not yield amplicons, mecB-specific PCR applying total and plasmid DNA resulted in a PCR product similar to those of M. caseolyticus isolate AM20CR01 (from a clam; isolate provided from J.E. Rubin, Institute of Veterinary Microbiology, University of Saskatchewan, Saskatchewan, Canada). Comparative analysis of the DNA sequence revealed complete sequence identity (100%) with reported mecB genes located either on plasmid pMCCL2 (M. caseolyticus JCSC5402; GenBank accession no. NC_011996.1) or within the SCCmec-like element of M. caseolyticus JCSC7096 (accession no. AB498756.1) (15,16). Apart from other mecB database entries, the highest nucleotide identity was shared with the sequence of mecD (68.7%), whereas the reported allotypes of mecC and mecA were more distantly related (Technical Appendix Figure 1). WGS revealed that the UKM4229 genome consists of a 2,851,374-bp circular chromosome and 2 different plasmids, a 20,725-bp plasmid (pSAWWU4229_2) and an 84,599-bp plasmid (pSAWWU4229_1; Figure); the latter carried mecB (GenBank accession no. PRJEB19527). The pSAWWU4229_1 plasmid backbone showed the highest similarity with the plasmid pMCLL2 of M. caseolyticus JCSC5402 (GenBank accession no. AP009486.1; blastn [https://blast.ncbi.nlm.nih.gov/Blast.cgi] 2.7.0+ maximum score 27,835; query coverage 71%; identity 99%) (33). These 2 plasmids shared 73.3% nucleotide identity (global alignment using Stretcher [Emboss], Matrix EDNAFULL; gap penalty 16, extend penalty 4). Whole plasmid comparative analysis of the sequences of pSAWWU4229_1 and pMCCL2 showed homologous regions between the mec gene complex, the downstream part of the mec complex, and the other antibiotic drug resistance genes (Technical Appendix Figure 2).
Within the pSAWWU4229_1 plasmid, mecB was flanked by β-lactam regulatory genes similar to mecR, mecI, and blaZ (nucleotide identities: 99.9%, mecRm from M. caseolyticus JCSC7096; 100%, mecIm from M. caseolyticus JCSC7096 and 100%, blaZm from M. caseolyticus JCSC7096). pSAWWU4229_1 contained additional antibiotic drug resistance genes encoding resistance to aminoglycosides (aacA-aphD, aphA, and aadK), as well as macrolides (ermB), tetracyclines (tetS), and streptothricin (sat), all located in the same gene section. This particular region of the plasmid showed similarities with the transposon Tn551 of S. aureus 4578 (Genbank accession no. LC125350.1; blastn 2.7.0+ maximum score 11,064; query coverage 10%; identity 99%) (34). The sequences shared 48.9% nucleotide identity (global alignment using Stretcher [Emboss], Matrix EDNAFULL; gap penalty 16, extend penalty 4). Mating-pore genes or genes responsible for the DNA transfer suggesting self-transmission or mobilization properties of pSAWWU4229_1 were not detected. Genotyping revealed that S. aureus isolate UKM4229 belonged to multilocus sequence typing type ST7 and spa-type t091 (spa-CC 091).
DNA microarray analysis and WGS revealed the isolate possessed the leucotoxin genes lukF, lukS, lukD, lukE, lukX, and lukY. The isolate belonged to capsule type 8, and the biofilm-associated genes icaA, C, and D were detected. Furthermore, the hlb-converting bacteriophage of immune-evasion cluster type G comprising the enterotoxin encoding genes sep, sak, and scn was present in the genome. Additional information about the virulence profile of this isolate is given in Technical Appendix Table 2.
Recent studies have shown that mobile SCCmec elements have been imported more frequently by different S. aureus clonal lineages than previously assessed (35). Nevertheless, in contrast to the huge diversity of non-MRSA S. aureus clonal lineages (36), comparatively few clonal lineages still dominate the global MRSA population (37). However, an increased transferability of methicillin resistance by a plasmid-encoded course of action would have the capacity to drastically change the MRSA epidemiology. In staphylococci and other members of the phylum Firmicutes, plasmids have contributed enormously to the emergence and spread of antimicrobial resistance, and plasmid-encoded penicillin resistance has reached or exceeded 80% of clinical staphylococcal isolates (38).
In M. caseolyticus, mecB genes have been found within the chromosome as part of an SCCmec element as well as on a plasmid (15,16,37). For S. aureus UKM4229, it was shown that the mecB carrying plasmid pSAWWU4229_1 was distantly related to a macrococcal plasmid (pMCLL2 of M. caseolyticus JCSC5402), substantiating a possible gene transfer between the two genera. Because macrococcal and staphylococcal species may share the same hosts, mammalian skin and food, an exchange of mobile genetic elements between members of both closely related genera is likely and transmission to mammal-adapted staphylococci is generally to be feared (3). Genotyping of S. aureus UKM4229 revealed spa-type t091, which is relatively common, as 0.92% of the >370,000 submitted spa sequences assigned to ≈17,000 spa types (as of February 2017) of the RIDOM SpaServer database (http://spa.ridom.de/spatypes.shtml) belong to this spa type.
Routine phenotypic methods for susceptibility testing cannot distinguish between methicillin resistance determinants; thus, mecB-encoded methicillin resistance can remain undiscovered. Moreover, mecB detection is not part of molecular screening approaches. Certain clonal lineages of S. aureus, including MRSA, have emerged as zoonotic pathogens colonizing farm and wild animals (40). Tetracycline resistance frequently observed in staphylococci associated with husbandry is another indication for a possible livestock origin of the isolate (41). A putative livestock source of the mecB-encoding plasmid underlines the importance of the One Health concept in combating the spread of antimicrobial drug resistance.
Although the mecB isolate has been tested susceptible toward several agents of non–β- lactam antibiotic drug classes, the generally increased risk, compared to that of a SCCmec transfer, should be taken into consideration in that a mecB-encoding plasmid will be transmitted through horizontal gene transfer to other staphylococcal strains, even to already multidrug-resistant strains. In S. aureus, 2 major means of horizontal gene transfer for plasmids have been described: conjugation and bacteriophage transduction. Here, pSAWWU4229_1 did not harbor the typical genes responsible for conjugation or mobilization, which is, however, a common lack in S. aureus, affecting ≈95% of plasmids (412 In contrast, for most staphylococcal plasmids, a transfer through bacteriophage generalized transduction has been suggested (43,44). Further studies are warranted to underpin this putative threat and to investigate how a plasmidborne methicillin resistance would affect the SCCmec-based methicillin resistance. For UKM4229, the WGS data revealed that the SCCmec chromosomal attachment site (attB) locus and the neighboring orfX (rlmH) gene were intact, and no integration of an SCCmec element was found.
The mecB isolate was tested to be susceptible to ceftobiprole and ceftaroline. Although cephalosporins with anti-MRSA activity are still active against the majority of MRSA isolates, nonsusceptibility has been already associated with certain MRSA lineages ranging between 3.9% and 33.5% of all MRSA isolates (45–48).
The discovery of plasmid-encoded methicillin resistance in S. aureus of probably macrococcal origin in a healthcare setting reveals a novel level of risk of the transfer of broad β-lactam resistance in staphylococci. Further studies are needed to clarify the real prevalence of mecB-caused methicillin resistance among MRSA and methicillin-resistant coagulase-negative staphylococci in human and animal populations, whether mecA and mecC genes could be found integrated on plasmids, and how the answers to these questions may affect human and animal health.
Dr. Becker is a professor of medical microbiology at the Institute of Medical Microbiology, University Hospital Münster, Münster, Germany. His research is focused on the epidemiology, pathogenesis, diagnosis, prevention, and therapy of staphylococcal infections. In particular, he has done extensive research on the characterization of MRSA and the staphylococcal small colony-variant phenotype.
Acknowledgments
We thank the Technology Development group, in particular Robert Sebra from the Icahn Institute for Genomics and Multiscale Biology at the Icahn School of Medicine at Mount Sinai, for PacBio sequencing and assembly, as well as Christian Ruckert from the Institute of Genomics at the University Hospital of Münster for bioinformatics support. We thank the GenDB support team for technical assistance and access to resources financially supported by the German Federal Ministry of Education and Research (BMBF) (FKZ 031A533) within the de.NBI network. Furthermore, we thank Jörg Wüllenweber for supervising the routine diagnostics leading to the isolation and presumptive identification of the isolate, as well as Melanie Bach and Martina Schulte for excellent technical assistance.
This work was supported in part by the BMBF within the frameworks of the Infect Control 2020 consortium [03ZZ0802H to K.B. and G.P.], #1Health-PREVENT (01KI1727A to K.B. and A.M.) and the German Center for Infection Research TTU 08.807 (8037808809 to K.B. and G.P.); and by the European Regional Development Fund within the EurHealth-1Health project (EU/INTERREG VA-681377 to K.B.).
References
- de Kraker ME, Davey PG, Grundmann H; BURDEN study group. Mortality and hospital stay associated with resistant Staphylococcus aureus and Escherichia coli bacteremia: estimating the burden of antibiotic resistance in Europe. PLoS Med. 2011;8:e1001104. DOIPubMedGoogle Scholar
- Lee BY, Singh A, David MZ, Bartsch SM, Slayton RB, Huang SS, et al. The economic burden of community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA). Clin Microbiol Infect. 2013;19:528–36. DOIPubMedGoogle Scholar
- Becker K, Heilmann C, Peters G. Coagulase-negative staphylococci. Clin Microbiol Rev. 2014;27:870–926. DOIPubMedGoogle Scholar
- Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG Jr. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. 2015;28:603–61. DOIPubMedGoogle Scholar
- Johnson AP. Methicillin-resistant Staphylococcus aureus: the European landscape. J Antimicrob Chemother. 2011;66(Suppl 4):iv43–8. DOIPubMedGoogle Scholar
- Chambers HF, Deleo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol. 2009;7:629–41. DOIPubMedGoogle Scholar
- Harris SR, Feil EJ, Holden MT, Quail MA, Nickerson EK, Chantratita N, et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327:469–74. DOIPubMedGoogle Scholar
- Becker K, Ballhausen B, Kahl BC, Köck R. The clinical impact of livestock-associated methicillin-resistant Staphylococcus aureus of the clonal complex 398 for humans. Vet Microbiol. 2017;200:33–8. DOIPubMedGoogle Scholar
- Peacock SJ, Paterson GK. Mechanisms of methicillin resistance in Staphylococcus aureus. Annu Rev Biochem. 2015;84:577–601. DOIPubMedGoogle Scholar
- Becker K, Ballhausen B, Köck R, Kriegeskorte A. Methicillin resistance in Staphylococcus isolates: the “mec alphabet” with specific consideration of mecC, a mec homolog associated with zoonotic S. aureus lineages. Int J Med Microbiol. 2014;304:794–804. DOIPubMedGoogle Scholar
- Shore AC, Deasy EC, Slickers P, Brennan G, O’Connell B, Monecke S, et al. Detection of staphylococcal cassette chromosome mec type XI carrying highly divergent mecA, mecI, mecR1, blaZ, and ccr genes in human clinical isolates of clonal complex 130 methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2011;55:3765–73. DOIPubMedGoogle Scholar
- García-Álvarez L, Holden MT, Lindsay H, Webb CR, Brown DF, Curran MD, et al. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect Dis. 2011;11:595–603. DOIPubMedGoogle Scholar
- Beck WD, Berger-Bächi B, Kayser FH. Additional DNA in methicillin-resistant Staphylococcus aureus and molecular cloning of mec-specific DNA. J Bacteriol. 1986;165:373–8. DOIPubMedGoogle Scholar
- Baba T, Kuwahara-Arai K, Uchiyama I, Takeuchi F, Ito T, Hiramatsu K. Complete genome sequence of Macrococcus caseolyticus strain JCSCS5402, [corrected] reflecting the ancestral genome of the human-pathogenic staphylococci. J Bacteriol. 2009;191:1180–90. DOIPubMedGoogle Scholar
- Tsubakishita S, Kuwahara-Arai K, Baba T, Hiramatsu K. Staphylococcal cassette chromosome mec-like element in Macrococcus caseolyticus. Antimicrob Agents Chemother. 2010;54:1469–75. DOIPubMedGoogle Scholar
- Schwendener S, Cotting K, Perreten V. Novel methicillin resistance gene mecD in clinical Macrococcus caseolyticus strains from bovine and canine sources. Sci Rep. 2017;7:43797. DOIPubMedGoogle Scholar
- Köck R, Werner P, Friedrich AW, Fegeler C, Becker K, Bindewald O, et al.; Prevalence of Multiresistant Microorganisms (PMM) Study Group; Prevalence of Multiresistant Microorganisms PMM Study Group. Persistence of nasal colonization with human pathogenic bacteria and associated antimicrobial resistance in the German general population. New Microbes New Infect. 2015;9:24–34. DOIPubMedGoogle Scholar
- Lowy FD. Antimicrobial resistance: the example of Staphylococcus aureus. J Clin Invest. 2003;111:1265–73. DOIPubMedGoogle Scholar
- Stojanov M, Moreillon P, Sakwinska O. Excision of staphylococcal cassette chromosome mec in methicillin-resistant Staphylococcus aureus assessed by quantitative PCR. BMC Res Notes. 2015;8:828. DOIPubMedGoogle Scholar
- Borg MA, Hulscher M, Scicluna EA, Richards J, Azanowsky JM, Xuereb D, et al. Prevention of meticillin-resistant Staphylococcus aureus bloodstream infections in European hospitals: moving beyond policies. J Hosp Infect. 2014;87:203–11. DOIPubMedGoogle Scholar
- Jurke A, Köck R, Becker K, Thole S, Hendrix R, Rossen J, et al. Reduction of the nosocomial meticillin-resistant Staphylococcus aureus incidence density by a region-wide search and follow-strategy in forty German hospitals of the EUREGIO, 2009 to 2011. Euro Surveill. 2013;18:20579. DOIPubMedGoogle Scholar
- Liu P, Wu Z, Xue H, Zhao X. Antibiotics trigger initiation of SCCmec transfer by inducing SOS responses. Nucleic Acids Res. 2017;45:3944–52. DOIPubMedGoogle Scholar
- Humphreys H, Becker K, Dohmen PM, Petrosillo N, Spencer M, van Rijen M, et al. Staphylococcus aureus and surgical site infections: benefits of screening and decolonization before surgery. J Hosp Infect. 2016;94:295–304. DOIPubMedGoogle Scholar
- Mellmann A, Friedrich AW, Rosenkötter N, Rothgänger J, Karch H, Reintjes R, et al. Automated DNA sequence-based early warning system for the detection of methicillin-resistant Staphylococcus aureus outbreaks. PLoS Med. 2006;3:e33. DOIPubMedGoogle Scholar
- Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–15.PubMedGoogle Scholar
- Becker K, Pagnier I, Schuhen B, Wenzelburger F, Friedrich AW, Kipp F, et al. Does nasal cocolonization by methicillin-resistant coagulase-negative staphylococci and methicillin-susceptible Staphylococcus aureus strains occur frequently enough to represent a risk of false-positive methicillin-resistant S. aureus determinations by molecular methods? J Clin Microbiol. 2006;44:229–31. DOIPubMedGoogle Scholar
- Kriegeskorte A, Ballhausen B, Idelevich EA, Köck R, Friedrich AW, Karch H, et al. Human MRSA isolates with novel genetic homolog, Germany. Emerg Infect Dis. 2012;18:1016–8. DOIPubMedGoogle Scholar
- Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27:1009–10. DOIPubMedGoogle Scholar
- Peters G, Becker K. Epidemiology, control and treatment of methicillin-resistant Staphylococcus aureus. Drugs. 1996;52(Suppl 2):50–4. DOIPubMedGoogle Scholar
- Idelevich EA, Schaumburg F, Knaack D, Scherzinger AS, Mutter W, Peters G, et al. The recombinant bacteriophage endolysin HY-133 exhibits in vitro activity against different african clonal lineages of the Staphylococcus aureus complex, including Staphylococcus schweitzeri. Antimicrob Agents Chemother. 2016;60:2551–3. DOIPubMedGoogle Scholar
- Idelevich EA, von Eiff C, Friedrich AW, Iannelli D, Xia G, Peters G, et al. In vitro activity against Staphylococcus aureus of a novel antimicrobial agent, PRF-119, a recombinant chimeric bacteriophage endolysin. Antimicrob Agents Chemother. 2011;55:4416–9. DOIPubMedGoogle Scholar
- Meyer F, Goesmann A, McHardy AC, Bartels D, Bekel T, Clausen J, et al. GenDB—an open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 2003;31:2187–95. DOIPubMedGoogle Scholar
- Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–14. DOIPubMedGoogle Scholar
- Nübel U, Roumagnac P, Feldkamp M, Song JH, Ko KS, Huang YC, et al. Frequent emergence and limited geographic dispersal of methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci U S A. 2008;105:14130–5. DOIPubMedGoogle Scholar
- Becker K, Schaumburg F, Fegeler C, Friedrich AW, Köck R; Prevalence of Multiresistant Microorganisms PMM Study. Staphylococcus aureus from the German general population is highly diverse. Int J Med Microbiol. 2017;307:21–7. DOIPubMedGoogle Scholar
- Monecke S, Coombs G, Shore AC, Coleman DC, Akpaka P, Borg M, et al. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS One. 2011;6:e17936. DOIPubMedGoogle Scholar
- Lanza VF, Tedim AP, Martínez JL, Baquero F, Coque TM. The plasmidome of Firmicutes: impact on the emergence and the spread of resistance to antimicrobials. Microbiol Spectr. 2015;3:PLAS-0039–2014.
- Gómez-Sanz E, Schwendener S, Thomann A, Gobeli Brawand S, Perreten V. First staphylococcal cassette chromosome mec containing a mecB-carrying gene complex independent of transposon Tn6045 in a Macrococcus caseolyticus isolate from a canine infection. Antimicrob Agents Chemother. 2015;59:4577–83. DOIPubMedGoogle Scholar
- Harrison EM, Paterson GK, Holden MT, Larsen J, Stegger M, Larsen AR, et al. Whole genome sequencing identifies zoonotic transmission of MRSA isolates with the novel mecA homologue mecC. EMBO Mol Med. 2013;5:509–15. DOIPubMedGoogle Scholar
- Larsen J, Clasen J, Hansen JE, Paulander W, Petersen A, Larsen AR, et al. Copresence of tet(K) and tet(M) in livestock-associated methicillin-resistant Staphylococcus aureus clonal complex 398 is associated with increased fitness during exposure to sublethal concentrations of tetracycline. Antimicrob Agents Chemother. 2016;60:4401–3. DOIPubMedGoogle Scholar
- Ramsay JP, Kwong SM, Murphy RJ, Yui Eto K, Price KJ, Nguyen QT, et al. An updated view of plasmid conjugation and mobilization in Staphylococcus. Mob Genet Elements. 2016;6:e1208317. DOIPubMedGoogle Scholar
- McCarthy AJ, Lindsay JA. The distribution of plasmids that carry virulence and resistance genes in Staphylococcus aureus is lineage associated. BMC Microbiol. 2012;12:104. DOIPubMedGoogle Scholar
- Shearer JE, Wireman J, Hostetler J, Forberger H, Borman J, Gill J, et al. Major families of multiresistant plasmids from geographically and epidemiologically diverse staphylococci. G3 (Bethesda). 2011;1:581–91. DOIPubMedGoogle Scholar
- Chan LC, Basuino L, Diep B, Hamilton S, Chatterjee SS, Chambers HF. Ceftobiprole- and ceftaroline-resistant methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2015;59:2960–3. DOIPubMedGoogle Scholar
- Schaumburg F, Peters G, Alabi A, Becker K, Idelevich EA. Missense mutations of PBP2a are associated with reduced susceptibility to ceftaroline and ceftobiprole in African MRSA. J Antimicrob Chemother. 2016;71:41–4. DOIPubMedGoogle Scholar
- Farrell DJ, Castanheira M, Mendes RE, Sader HS, Jones RN. In vitro activity of ceftaroline against multidrug-resistant Staphylococcus aureus and Streptococcus pneumoniae: a review of published studies and the AWARE Surveillance Program (2008-2010). Clin Infect Dis. 2012;55(Suppl 3):S206–14. DOIPubMedGoogle Scholar
- Zhang H, Xiao M, Kong F, O’Sullivan MV, Mao LL, Zhao HR, et al. A multicentre study of meticillin-resistant Staphylococcus aureus in acute bacterial skin and skin-structure infections in China: susceptibility to ceftaroline and molecular epidemiology. Int J Antimicrob Agents. 2015;45:347–50. DOIPubMedGoogle Scholar
- Murakami K, Minamide W, Wada K, Nakamura E, Teraoka H, Watanabe S. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J Clin Microbiol. 1991;29:2240–4.PubMedGoogle Scholar
Figure
Tables
Cite This ArticleTable of Contents – Volume 24, Number 2—February 2018
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Karsten Becker, University Hospital Münster—Institute of Medical Microbiology, Domagkstr. 10, D-48149 Münster, Germany
Top