Volume 24, Number 5—May 2018
Research Letter
Borrelia miyamotoi sensu lato in Père David Deer and Haemaphysalis longicornis Ticks
Figure
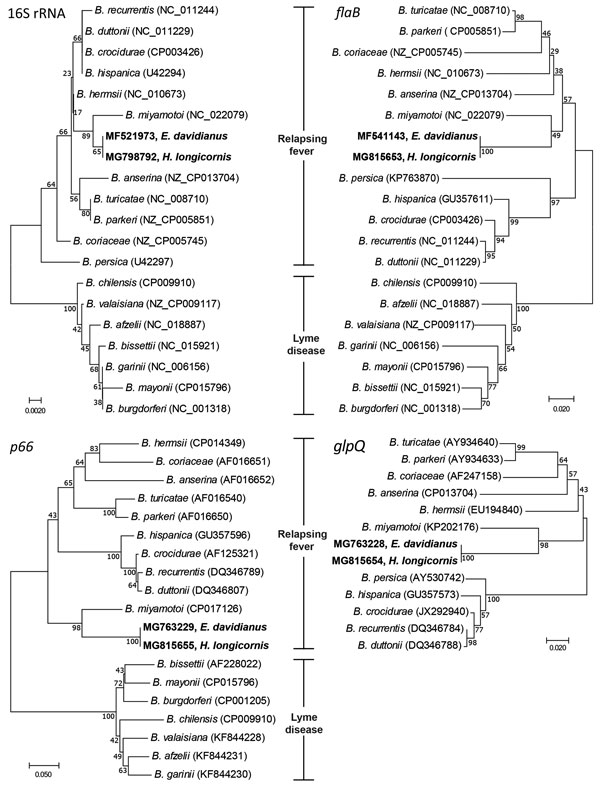
Figure. Neighbor-joining phylogenetic trees constructed with 16S rRNA, flaB, p66, and glpQ gene sequences of Borrelia spp. isolates collected from Père David deer (Elaphurus davidianus) and Haemaphysalis longicornis ticks, Dafeng Elk National Natural Reserve, China, and reference isolates. The isolates identified in this study (bold; GenBank accession nos. MF521973, MF541143, MG763228, MG763229) are most similar to B. miyamotoi of the relapsing fever group. Numbers at branch nodes show bootstrap support (1,000 replicates). Scale bars indicate nucleotide substitutions per site.
1These senior authors contributed equally to this article.
Page created: April 17, 2018
Page updated: April 17, 2018
Page reviewed: April 17, 2018
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.