Volume 24, Number 6—June 2018
Dispatch
Novel Salmonella enterica Serovar Typhimurium Genotype Levels as Herald of Seasonal Salmonellosis Epidemics
Cite This Article
Citation for Media
Abstract
We examined the population dynamics of Salmonella enterica serovar Typhimurium during seasonal salmonellosis epidemics in New South Wales, Australia, during 2009–2016. Of 15,626 isolates, 5%–20% consisted of novel genotypes. Seasons with salmonellosis epidemics were associated with a reduction in novel genotypes in the preceding winter and spring.
Nontyphoidal Salmonella spp. cause an estimated 93.8 million salmonellosis infections and 155,000 deaths globally each year (1). However, the population dynamics of human salmonellosis remain poorly understood, which can undermine the effective use of public health resources (2). Salmonella enterica serovar Typhimurium is a highly diverse serovar and the dominant cause of salmonellosis worldwide (3,4), experiencing continuous evolution, persistence, and adaptation within different ecologic niches. Whereas the complexities of the clonal structure of Salmonella Typhimurium populations have been recognized (4–7), the effect of temporal change in subtype diversity on disease incidence is not well understood.
Multilocus variable-number tandem-repeat (VNTR) analysis (MLVA) has been used as a high-resolution Salmonella typing method amenable to harmonization (8,9). In our study, we sought to determine if Salmonella Typhimurium subtype diversity can be used to predict incidence of human salmonellosis. We examined Salmonella Typhimurium isolates recovered during 2009–2016 in the comparatively low prevalence setting of New South Wales (NSW), the most populous state of Australia (Technical Appendix Table).
We used MLVA to genotype all Salmonella Typhimurium isolates referred to the NSW Enteric Reference Laboratory at the Centre for Infectious Diseases and Microbiology, NSW Health Pathology (Sydney, NSW, Australia), during August 2009–March 2016. We conducted multiplex PCR to amplify VNTRs (STTR9, STTR5, STTR6, STTR10pl and STTR3) and subsequent analyses as described previously (9,10). We reported MLVA results as a string of 5 numbers representing relevant repeats (11). We used Salmonella Typhimurium reference strain LT2 (GenBank accession on. NC_003197) as a control throughout and consistently generated the expected MLVA type 4-13-13-10-0211. We defined a cluster as >2 isolates with the same MLVA type collected within 12 weeks (10) and used a χ2 test to determine the significance of differences observed; we considered a p value of <0.05 significant. We determined population diversity by calculating the Simpson index of diversity (12), and population richness using the McIntosh dominance index (13). This study was approved by our local Human Research Ethics Committee (LNR/17/WMED/25).
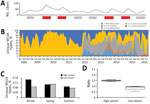
After excluding duplicate isolates from the same episode of the disease (99% of all cases recorded in NSW), we examined a total of 15,626 human Salmonella Typhimurium isolates. We defined seasons as spring (September–November), summer (December–February), autumn (March–May), and winter (June–August). We observed a substantial fluctuation in the number of infections during the study period (Figure 1). The relative contributions of common definitive phage types (DT) and MLVA types to local Salmonella Typhimurium activity varied over 8 seasonal peaks. DT135 dominated in 2008 but was subsequently replaced by DT170 as the most common type in 2009 and 2010; an increase in the activity of DT9 occurred in 2014 and 2015. In total, we observed 667 different MLVA types. Six related STM DT170 MLVA types (2-7-6-12-0212; 2-7-7-12-0212; 2-7-6-13-0212; 2-7-6-11-0212; 2-7-6-14-0212 and 2-7-7-11-0212) represented 30% of all isolates. Increases in Salmonella Typhimurium cases during summer and autumn months (December–May in the Southern Hemisphere), with 2 cycles of 2009–2011 and 2012–2014, were mirrored by an increase in the number of unique MLVA types detected. The proportions isolates included in clusters also demonstrated expected seasonal fluctuations corresponding to increases in incidence (Figure 1, panel A), although we found no significant change in the number of clusters or their average size during the study period (p<0.05).
The average number of Salmonella Typhimurium cases for the 8 summer–autumn seasons investigated during the study period was 1,301. We categorized all summer–autumn seasons as either high (epidemic) or low on the basis of whether case numbers were above or below the average. We confirmed the designations by comparing the seasonal averages calculated for this study with yearly rates of salmonellosis notifications. These 2 approaches congruently assigned summer–autumn seasons of 2010, 2011, 2014, and 2015 as high or epidemic, whereas summer–autumn seasons of 2009, 2012, 2013, and 2016 were classified as low; the difference between case numbers in high and low seasons was significant (p<0.02) (Figure 1, panels B, C). The average annual number of foodborne community outbreaks recorded by the national public health network during high seasons was 63 and during low seasons was 48 (http://www.ozfoodnet.gov.au).
At the beginning of the study, we considered all MLVA types novel. The ratio of novel, previously unreported MLVA types to all types stabilized within 5 months; after the fifth month, 10%–40% of all MLVA types detected at any given time consisted of novel MLVA types (Figure 2). We detected no changes in the age distribution of human populations affected by dominant types; 41.1% of all infections occurred in those <14 years of age.
The diversity of the Salmonella Typhimurium population remained relatively constant over time; the McIntosh dominance index of diversity fluctuated between 0.6 and 0.9 during both high and low seasons (p = 0.478). However, we observed a rapid decrease in the proportions of unique MLVA types from winter to spring (i.e., Uw/Usp ratio<1) before epidemic Salmonella Typhimurium activity. In contrast, the proportion of unique MLVA types increased from winter to spring preceding low seasons (Uw/Usp ratio>1) (Figure 2). This ratio also correlated with incidence of Salmonella Typhimurium in NSW over the study period (r = 0.922). Of note, the percentage of unique MLVA types recovered from patients <14 years of age during winter of high/epidemic seasons was also significantly lower than during the winter months of low seasons of salmonellosis (15.1% vs. 28.2%; p<0.001).
Our observations potentially reflect the decrease in Salmonella Typhimurium diversity resulting from the reduction of genome variation under selection pressure, which is associated with the emergence of successful unique clones capable of causing epidemics in immunologically naive hosts. However, longer-term monitoring of subtype diversity and disease incidence is warranted to confirm these trends. Although an analogous selection-driven reduction in genetic diversity has been observed in other pathogens, such as influenza virus (14), it has not previously been observed in Salmonella. Our findings also suggest that the timely recognition of novel Salmonella Typhimurium subtypes may be of significance for surveillance and that the conventional diversity indices alone may not be sufficient to detect subtle changes in circulating subtypes. As the estimated ratio of accumulation of MLVA repeats in different loci to single nucleotide polymorphisms is 1:6.9 (15), changes in the composition of Salmonella Typhimurium subtypes might offer insight into the relevance of population diversity for fluctuations in the incidence of salmonellosis.
Substantial increases in seasonal epidemics of Salmonella Typhimurium can be associated with a reduction in newly identified MLVA types in the preceding winter and spring, reflecting the emergence of successful Salmonella Typhimurium clones under selection pressure. The proportion of novel MLVA types in winter and spring may serve as an early warning sign in public health surveillance. These observations add further insights into the epidemiology of Salmonella Typhimurium infections in a low-incidence setting. Although they may not be readily applicable to high-incidence Salmonella Typhimurium settings with frequent co-infections and different diagnostic or public health practices, the epidemiology of Salmonella Typhimurium and public health responses in Australia are similar to those in other industrialized countries, supporting the generalizability of our findings. Prospective monitoring of Salmonella Typhimurium population diversity and identifying new MLVA types as reservoirs from which future epidemics might emerge can improve the assessment of risks of seasonal increase in Salmonella Typhimurium incidence.
Dr. Sotomayor is a veterinary scientist and a postgraduate researcher with Sydney Medical School, The University of Sydney, Sydney, New South Wales, Australia. She applies molecular subtyping methods to investigate mechanisms of epidemics of foodborne salmonellosis.
Acknowledgments
We thank the Microbiological Diagnostic Unit of the University of Melbourne for providing phage typing data and the public health epidemiologists from the Health Protection Branch of New South Wales Ministry of Health and OzFoodNet.
This study was funded in part by the NSW Department of Health by its Capacity Building Grant. C.S. was supported by a Becas Chile Scholarship from the Ministry of Education of Chile and V.S. was funded by the Australian National Health & Medical Council (grant 457122).
References
- Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O’Brien SJ, et al.; International Collaboration on Enteric Disease ‘Burden of Illness’ Studies. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis. 2010;50:882–9. DOIPubMedGoogle Scholar
- Kirk MD, Pires SM, Black RE, Caipo M, Crump JA, Devleesschauwer B, et al. World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal and viral diseases, 2010: a data synthesis. [Erratum in: PLoS Med 2015;12:e1001940.]. PLoS Med. 2015;12:e1001921. DOIPubMedGoogle Scholar
- Ford L, Glass K, Veitch M, Wardell R, Polkinghorne B, Dobbins T, et al. Increasing incidence of Salmonella in Australia, 2000–2013. PLoS One. 2016;11:e0163989. DOIPubMedGoogle Scholar
- Ao TT, Feasey NA, Gordon MA, Keddy KH, Angulo FJ, Crump JA. Global burden of invasive nontyphoidal Salmonella disease, 2010(1). Emerg Infect Dis. 2015;21:941–9. DOIPubMedGoogle Scholar
- Lan R, Reeves PR, Octavia S. Population structure, origins and evolution of major Salmonella enterica clones. Infect Genet Evol. 2009;9:996–1005. DOIPubMedGoogle Scholar
- Petrovska L, Mather AE, AbuOun M, Branchu P, Harris SR, Connor T, et al. Microevolution of monophasic Salmonella Typhimurium during epidemic, United Kingdom, 2005–2010. Emerg Infect Dis. 2016;22:617–24. DOIPubMedGoogle Scholar
- Langridge GC, Fookes M, Connor TR, Feltwell T, Feasey N, Parsons BN, et al. Patterns of genome evolution that have accompanied host adaptation in Salmonella. Proc Natl Acad Sci U S A. 2015;112:863–8. DOIPubMedGoogle Scholar
- Lindstedt B-A, Vardund T, Aas L, Kapperud G. Multiple-locus variable-number tandem-repeats analysis of Salmonella enterica subsp. enterica serovar Typhimurium using PCR multiplexing and multicolor capillary electrophoresis. J Microbiol Methods. 2004;59:163–72. DOIPubMedGoogle Scholar
- Torpdahl M, Sørensen G, Lindstedt BA, Nielsen EM. Tandem repeat analysis for surveillance of human Salmonella Typhimurium infections. Emerg Infect Dis. 2007;13:388–95. DOIPubMedGoogle Scholar
- Sintchenko V, Wang Q, Howard P, Ha CWY, Kardamanidis K, Musto J, et al. Improving resolution of public health surveillance for human Salmonella enterica serovar Typhimurium infection: 3 years of prospective multiple-locus variable-number tandem-repeat analysis (MLVA). BMC Infect Dis. 2012;12:78. DOIPubMedGoogle Scholar
- Larsson JT, Torpdahl M, Petersen RF, Sørensen G, Lindstedt BA, Nielsen EM. Development of a new nomenclature for Salmonella typhimurium multilocus variable number of tandem repeats analysis (MLVA). Euro Surveill. 2009;14:19174.PubMedGoogle Scholar
- Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol. 1988;26:2465–6.PubMedGoogle Scholar
- Magurran AE. Ecological diversity and its measurement. London: Chapman & Hall; 1988.
- Bahl J, Nelson MI, Chan KH, Chen R, Vijaykrishna D, Halpin RA, et al. Temporally structured metapopulation dynamics and persistence of influenza A H3N2 virus in humans. Proc Natl Acad Sci U S A. 2011;108:19359–64. DOIPubMedGoogle Scholar
- Fu S, Octavia S, Wang Q, Tanaka MM, Tay CY, Sintchenko V, et al. Evolution of variable number tandem repeats and its relationship with genomic diversity in Salmonella Typhimurium. Front Microbiol. 2016;7:2002. DOIPubMedGoogle Scholar
Figures
Cite This ArticleOriginal Publication Date: May 02, 2018
Table of Contents – Volume 24, Number 6—June 2018
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Vitali Sintchenko, Centre for Infectious Diseases and Microbiology, Level 3, Institute of Clinical Pathology and Medical Research, Westmead Hospital, Westmead, Sydney, NSW 2145, Australia
Top