Volume 24, Number 6—June 2018
Research
Novel Parvovirus Related to Primate Bufaviruses in Dogs
Figure 2
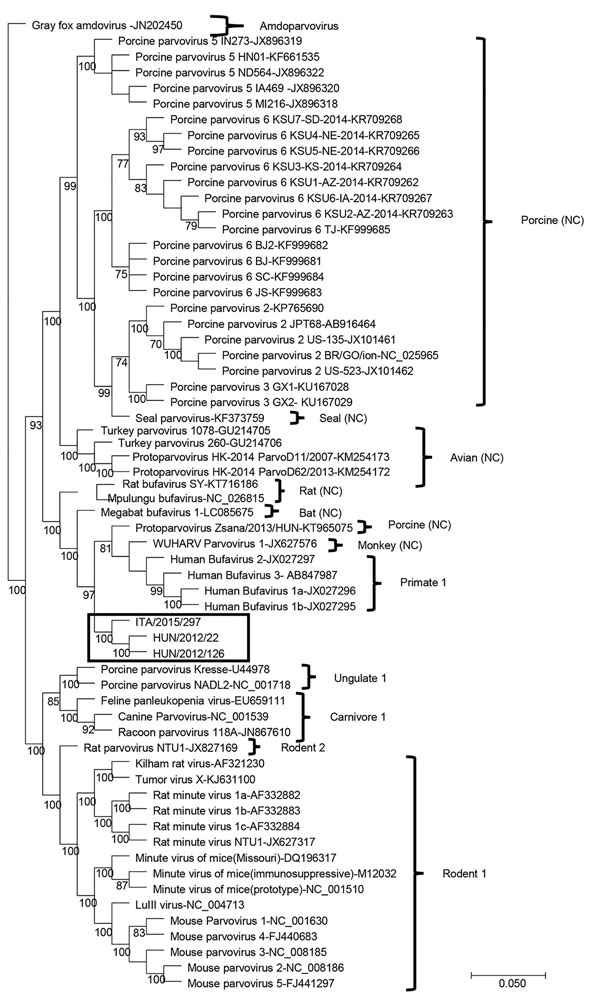
Figure 2. Capsid-based phylogenetic tree displaying the diversity of protoparvoviruses. The protoparvoviruses officially recognized by the International Committee on Taxonomy of Viruses are included, along with nonclassified (NC) protoparvoviruses. The tree was generated using the neighbor-joining method with the Jukes-Cantor algorithm of distance correction, with bootstrapping over 1,000 replicates. Box indicates canine bufavirus strains. GenBank accession numbers are provided for reference isolates; gray fox amdovirus (GenBank accession no. JN202450) is used as outgroup. Scale bar indicates nucleotide substitutions per site.
Page created: May 17, 2018
Page updated: May 17, 2018
Page reviewed: May 17, 2018
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.