Volume 24, Number 6—June 2018
Dispatch
Novel Poxvirus in Proliferative Lesions of Wild Rodents in East Central Texas, USA
Abstract
Northern pygmy mice from 2 localities in East Central Texas, USA, had proliferative epidermal lesions on the tail and feet. Electron microscopy of lesion tissue revealed poxvirus. Phylogenetic analyses indicated the virus differed 35% from its closest relatives, the Chordopoxvirinae. Future research is needed to determine whether this virus could affect human health.
Chordopoxvirinae is a diverse subfamily of viruses within Poxviridae. These geographically widespread viruses infect birds, reptiles, and mammals, and many are zoonotic (1). The increasing use of molecular methods has resulted in the identification of several novel poxviruses from humans and animals, many of which probably represent new genera (2–9); often, the reported host range of these viruses is limited to the species of the index case. Novel human poxvirus infections identified in the 21st century have often been presumed to have animal origins; for example, an investigation of a novel poxvirus isolated from 2 men in the country of Georgia revealed serologic evidence of orthopoxvirus exposure in cows in their herd and in captured rodents (5). We report a novel poxvirus infection characterized by proliferative epidermal lesions in wild northern pygmy mice (Baiomys taylori) found at 2 localities in east central Texas, USA, and further characterize the virus through genetic analysis.
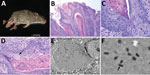
In August 2014, we captured a severely affected adult male B. taylori mouse (mouse 1) at the Attwater Prairie Chicken National Wildlife Refuge in Colorado County, Texas. The mouse had large (4–8-mm diameter) proliferative lesions on the hind feet and tail (Figure 1, panel A) but otherwise appeared healthy. In April 2017, at the Biodiversity Research and Teaching Collections at Texas A&M University in College Station, Texas (160 km north of the first locality), we captured an additional adult male B. taylori mouse (mouse 2) with mild 1–2-mm proliferative lesions on the left hind foot and tail. Both animals were euthanized in accordance with Texas Parks and Wildlife Department scientific collections permit (SPR-0512-917) and Texas A&M University Institutional Animal Care and Use Committee’s animal use protocol (2015-0088). These 2 B. taylori specimens are housed at Biodiversity Research and Teaching Collections (mammal voucher nos. TCWC 65223 and TCWC 65224; http://portal.vertnet.org/search).
We subjected mouse 1 to a full necropsy and found the mouse to be in good body condition. Extending from the skin of the dorsal aspect of the left hind foot, plantar aspect of the right hind foot, and the dorsal tail were several firm, pedunculated, irregular masses 0.4–0.8 cm in diameter (Figure 1, panel A). When the masses were sectioned, the cut surface was light tan with a papillated appearance, and the masses did not appear to invade the underlying tissues. The tail was partially amputated but was healed and apparently unrelated to the lesions. We did not observe any other lesions. We froze a section of a mass at −80°C for molecular work and fixed the remaining tissues in 10% neutral-buffered formalin; the fixed tissues were processed for histology and stained with hematoxylin and eosin.
On microscopic examination, necrotizing and proliferative dermatitis was observed. The epidermis of the affected area had multiple large, exophytic, pedunculated masses composed of markedly hyperplastic epithelial cells forming papillary projections with abundant orthokeratotic and parakeratotic hyperkeratosis (Figure 1, panel B). The epithelial surface was multifocally eroded to ulcerated, and the stratum corneum contained aggregates of coccoid bacteria. The stratum spinosum and stratum granulosum were markedly thickened with swollen keratinocytes (ballooning degeneration) frequently containing intracytoplasmic eosinophilic viral inclusions (Figure 1, panels B, C). These inclusions frequently extended extracellularly, forming large lakes <50 μm in diameter. The dermis and stratum basale were infiltrated by lymphocytes, plasma cells, and macrophages and a large number of viable and degenerate neutrophils. We examined sections of the spleen, liver, lungs, heart, kidneys, and intestines and identified no substantial lesions. A viral etiology was suspected, so formalin-fixed paraffin-embedded sections of the mass were processed for transmission electron microscopy. Ultra-thin sections were examined with a Morgagni 268 transmission electron microscope (FEI, Hillsboro, OR, USA) at an accelerating voltage of 80 kV. Cytoplasmic inclusions contained granular, electron-dense material and numerous brick-shaped virions with dense cores and inner and outer membrane shell consistent with poxvirus (Figure 1, panel D).
We extracted DNA from skin masses of both mouse 1 and mouse 2 and subjected the DNA to PCR with low–GC content poxvirus primers targeting a region of the putative metalloproteinase gene (10) and Sanger sequencing. The virus sequences (220 bp) from mouse 1 (BtTX2014) and mouse 2 (BtTX2017) were identical to each other (GenBank accession no. MG367479), and the top 10 matches to this sequence in GenBank were all poxviruses with only 76%–78% shared identity. In addition, we extracted DNA from formalin-fixed paraffin-embedded sections of liver, lung, kidney, and spleen from mouse 1, and all were PCR negative for poxvirus DNA. A DNA aliquot of the mass from mouse 1 was sent to Otogenetics (Atlanta, GA, USA) for whole-genome sequencing with an Illumina HiSeq platform (Illumina Inc., San Diego, CA, USA). From these data, we extracted 9 core genes of BtTX2014 located within the conserved coding portion of the poxvirus genome used in previous studies (5) (GenBank accession nos. MG367480–8). In Geneious version 8.1.4 (https://www.geneious.com/), we converted the gene sequences into amino acid sequences for the purpose of alignment to those of 49 other chordopoxviruses (Chordopoxvirinae) and 2 entomopoxviruses (Entomopxvirinae), which were used as outgroup taxa. We then concatenated the gene sequences (total alignment 27,674 bp) and conducted partitioned phylogenetic analyses using Bayesian approaches in MrBayes version 3.2.2 (http://mrbayes.sourceforge.net/download.php). The phylogenetic position of this virus revealed it to be a divergent member of the Chordopoxvirinae (Figure 2), divergent from other Chordopoxvirinae poxviruses by an average of 35% (uncorrected p distances for the 27,674 bp of the concatenated alignment; Technical Appendix Table).
The poxvirus sequenced from B. taylori in east central Texas is distinct from previously identified viruses, with genetic distances similar to those observed between genera (online Technical Appendix Table). Although support for several phylogenetic relationships is low (possibly due to high genetic variation within Chordopoxvirinae), the genetic data strongly suggest this poxvirus does not belong to any recognized genus as of March 2018. We propose the tentative species name Brazospox virus in reference to the proximity of both field sites to the Brazos River.
The epidermal lesions produced by the virus are unique among previously described poxviruses of wild rodents, being proliferative rather than ulcerative in nature, and lacking systemic involvement (7,11). The population-level implications of this poxvirus on hosts are unclear. The 2 mice in this study represent the spectrum of observed pathology, from severe (mouse 1) to mild (mouse 2). The virus was confirmed at 2 different localities, and field notes indicate similar lesions were observed in other rodent species. During 2013–2017 (12,13), among ≈1,800 rodents captured during field research in east central Texas, we documented proliferative lesions on the tail or feet of >17 individual rodents of 3 species (B. taylori, n = 12; Chaetodipus hispidus, n = 2; Sigmodon hispidus, n = 3). The combined distributional range of these 3 host species includes >20 US states and a large portion of Mexico (14).
Novel poxviruses identified in wildlife populations might be useful for the identification of threats to human and animal health. The description of these new viruses contributes to the study of viral diversity and pathogenesis. Propagation of the Brazospox virus in cell culture for use in infection studies, coupled with expanded field surveillance, examination of museum specimens, and full genome analysis could yield additional clues to the origin, pathogenesis, and potential host range of this novel poxvirus.
Dr. Hodo is a veterinary pathologist and postdoctoral associate at the College of Veterinary Medicine at Texas A&M University (College Station, Texas), with interests in wildlife pathology, disease ecology, and epidemiology.
Acknowledgments
The authors thank H. Ross Payne for his technical expertise with electron microscopy and imaging. We thank Adrian Castellanos, Jaime Rodriguez, and Katelyn Wagner for fieldwork assistance. We thank Rebecca Chester, Mike Morrow, and Terry Rossignol for allowing property access to Attwater Prairie Chicken National Wildlife Refuge and Whitni Davidson for processing samples.
This work was supported in part by the Texas A&M AgriLife Invasive Fire Ant Research and Management Project. Fellowship support was provided by a National Institutes of Health fellowship (no. 2T32OD011083-06 to C.L.H.).
References
- Emerson GL, Li Y, Frace MA, Olsen-Rasmussen MA, Khristova ML, Govil D, et al. The phylogenetics and ecology of the orthopoxviruses endemic to North America. PLoS One. 2009;4:e7666–7. DOIPubMedGoogle Scholar
- Goldsmith CS, Metcalfe MG, Li Y, Davidson WB, Wilkins K, Roess AA, et al. Emergence of previously unknown poxviruses. Microsc Microanal. 2016;22(S3):1184–5. DOIGoogle Scholar
- Smithson C, Meyer H, Gigante CM, Gao J, Zhao H, Batra D, et al. Two novel poxviruses with unusual genome rearrangements: NY_014 and Murmansk. Virus Genes. 2017;53:883–97. DOIPubMedGoogle Scholar
- Shchelkunov SN. An increasing danger of zoonotic orthopoxvirus infections. PLoS Pathog. 2013;9:e1003756–4. DOIPubMedGoogle Scholar
- Vora NM, Li Y, Geleishvili M, Emerson GL, Khmaladze E, Maghlakelidze G, et al. Human infection with a zoonotic orthopoxvirus in the country of Georgia. N Engl J Med. 2015;372:1223–30. DOIPubMedGoogle Scholar
- Emerson GL, Nordhausen R, Garner MM, Huckabee JR, Johnson S, Wohrle RD, et al. Novel poxvirus in big brown bats, northwestern United States. Emerg Infect Dis. 2013;19:1002–4. DOIPubMedGoogle Scholar
- Wibbelt G, Tausch SH, Dabrowski PW, Kershaw O, Nitsche A, Schrick L. Berlin squirrelpox virus, a new poxvirus in red squirrels, Berlin, Germany. Emerg Infect Dis. 2017;23:1726–9. DOIPubMedGoogle Scholar
- Goldberg TL, Chapman CA, Cameron K, Saj T, Karesh WB, Wolfe ND, et al. Serologic evidence for novel poxvirus in endangered red Colobus monkeys, Western Uganda. Emerg Infect Dis. 2008;14:801–3. DOIPubMedGoogle Scholar
- Cardeti G, Gruber CEM, Eleni C, Carletti F, Castilletti C, Manna G, et al. Fatal outbreak in Tonkean macaques caused by possibly novel orthopoxvirus, Italy, January 2015. Emerg Infect Dis. 2017;23:1941–9. DOIPubMedGoogle Scholar
- Li Y, Meyer H, Zhao H, Damon IK. GC content-based pan-pox universal PCR assays for poxvirus detection. J Clin Microbiol. 2010;48:268–76. DOIPubMedGoogle Scholar
- Bangari DS, Miller MA, Stevenson GW, Thacker HL, Sharma A, Mittal SK. Cutaneous and systemic poxviral disease in red (Tamiasciurus hudsonicus) and gray (Sciurus carolinensis) squirrels. Vet Pathol. 2009;46:667–72. DOIPubMedGoogle Scholar
- Rodriguez JE, Hamer SA, Castellanos AA, Light JE. Survey of a rodent and tick community in east-central Texas. Southeast Nat. 2015;14:415–24. DOIGoogle Scholar
- Castellanos AA, Medeiros MCI, Hamer GL, Morrow ME, Eubanks MD, Teel PD, et al. Decreased small mammal and on-host tick abundance in association with invasive red imported fire ants (Solenopsis invicta). Biol Lett. 2016;12:20160463–5. DOIPubMedGoogle Scholar
- International Union for Conservation of Nature. The IUCN red list of threatened species. 2017 [cited 2017 Nov 16]. http://www.iucnredlist.org/
Figures
Cite This ArticleOriginal Publication Date: May 07, 2018
Table of Contents – Volume 24, Number 6—June 2018
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Sarah A. Hamer, Department of Veterinary Integrative Biosciences, 4458 TAMU, College Station, TX, 77843-4458, USA
Top