Volume 24, Number 7—July 2018
Research
Reemergence of Reston ebolavirus in Cynomolgus Monkeys, the Philippines, 2015
Figure 3
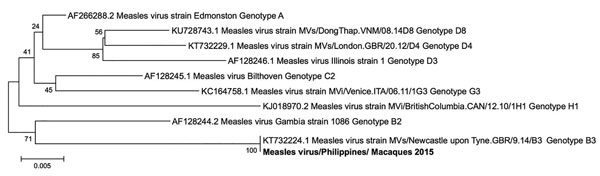
Figure 3. Phylogenetic tree (neighbor-joining) of the partial L gene (418 nt) of measles virus (GenBank accession no. MF496232) detected in macaques in 2015, produced by using MEGA 6 software (https://www.megasoftware.net). Numbers along branches indicate bootstrap values. Scale bar indicates nucleotide substitutions per site. Bold text indicates measles virus strain isolated in Philippines.
Page created: June 18, 2018
Page updated: June 18, 2018
Page reviewed: June 18, 2018
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.