Volume 25, Number 1—January 2019
Dispatch
Meat and Fish as Sources of Extended-Spectrum β-Lactamase–Producing Escherichia coli, Cambodia
Figure 2
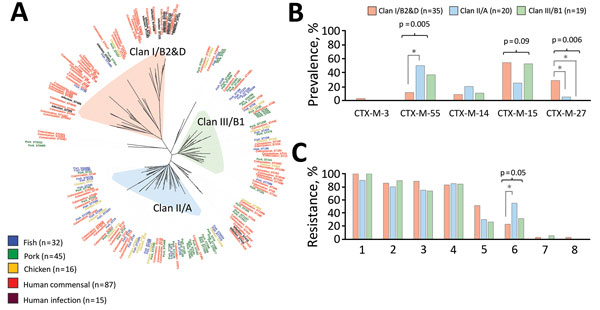
Figure 2. Genomic comparisons of extended-spectrum β-lactamase (ESBL)–producing Escherichia coli from humans, fish, pork, and chicken from Cambodia and differences in human colonization isolates by phylogenetic clan. All isolates were phenotypically resistant to third-generation cephalosporins (data not shown). A) Whole-genome sequence-based phylogenetic tree of 195 ESBL-producing E. coli genomes comprising 87 human colonization isolates, 15 human clinical isolates, and 93 isolates from fish, pork, and chicken meat and resulting phylogenetic clans I/B2&D (n = 53), II/A (n = 69), and III/B1 (n = 47). B) ESBL-encoding genes of human colonization E. coli isolates, by phylogenetic clan. C) Phenotypic resistance of human colonization ESBL-producing E. coli isolates to antimicrobial drugs of 8 classes, by phylogenetic clan. Clinical isolates are not included in panels B or C. Of 87 human colonization genomes, 13 did not group into a phylogenetic clan and thus are excluded from panels B and C. Prevalence of outcome differed significantly (p<0.05, indicated by *) between 2 indicated clans by post hoc Tukey test. Only statistically significant differences are depicted. 1, quinolone; 2, co-trimoxazole; 3, tetracycline; 4, aminoglycoside; 5, macrolide; 6, amphenicol; 7, carbapenem; 8, colistin.
1These senior authors contributed equally to this article.
2Additional members of the BIRDY study group are listed at the end of this article.