Volume 25, Number 10—October 2019
Dispatch
Antigenic Variation of Avian Influenza A(H5N6) Viruses, Guangdong Province, China, 2014–2018
Figure 2
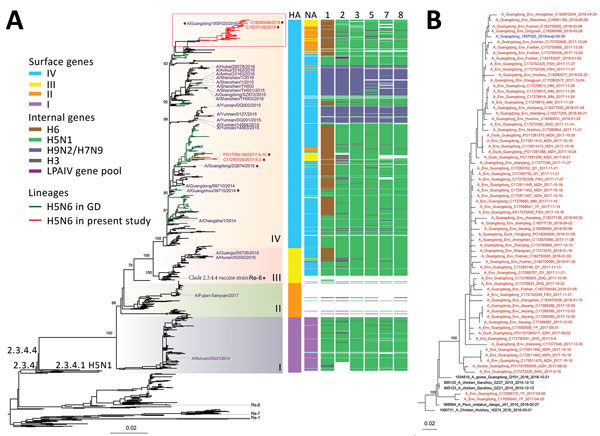
Figure 2. Phylogeny of influenza A(H5N6) viruses collected in Guangdong Province, China, January 2013–October 2018, compared with reference isolates. A) Viruses of clade 2.3.4.4 H5N6 viruses are divided into 4 subgroups (I–V) on the basis of the surface genes (HA and NA). Colors in key distinguish surface and internal genes. The A/chicken/Guizhou/4/2013 (Re-8) vaccine strain and viral strains used for HI testing are labeled. The 2018 human H5N6 isolate from Guangdong Province is blue, human H5N6 virus sequences since 2013 are purple, human and environmental H5N6 isolates used for the HI test are labeled with a purple dot (except for HA256 human strain, for which no sequence was available). The top part of the tree containing the bulk of the Guangdong Province recent H5N6 viruses and the human case is highlighted with a red box. All branch lengths are scaled according to the number of substitutions per site. Scale bar indicates nucleotide substitutions per site. GD, Guangdong; HA, hemagglutinin gene; LPAIV, low pathogenicity avian influenza virus; NA, neuraminidase gene; 1, polymerase basic 2 gene; 2, polymerase basic 1 gene; 3, polymerase acidic gene; 5, nucleoprotein gene; 7, matrix gene; 8, nonstructural gene. B) An expansion of the phylogenetic tree in the red outlined box of panel A. The sequence in blue is the newly approved vaccine strain 18SF020.
1These authors contributed equally to this article.