Volume 25, Number 10—October 2019
Dispatch
Plasmodium cynomolgi as Cause of Malaria in Tourist to Southeast Asia, 2018
Figure 2
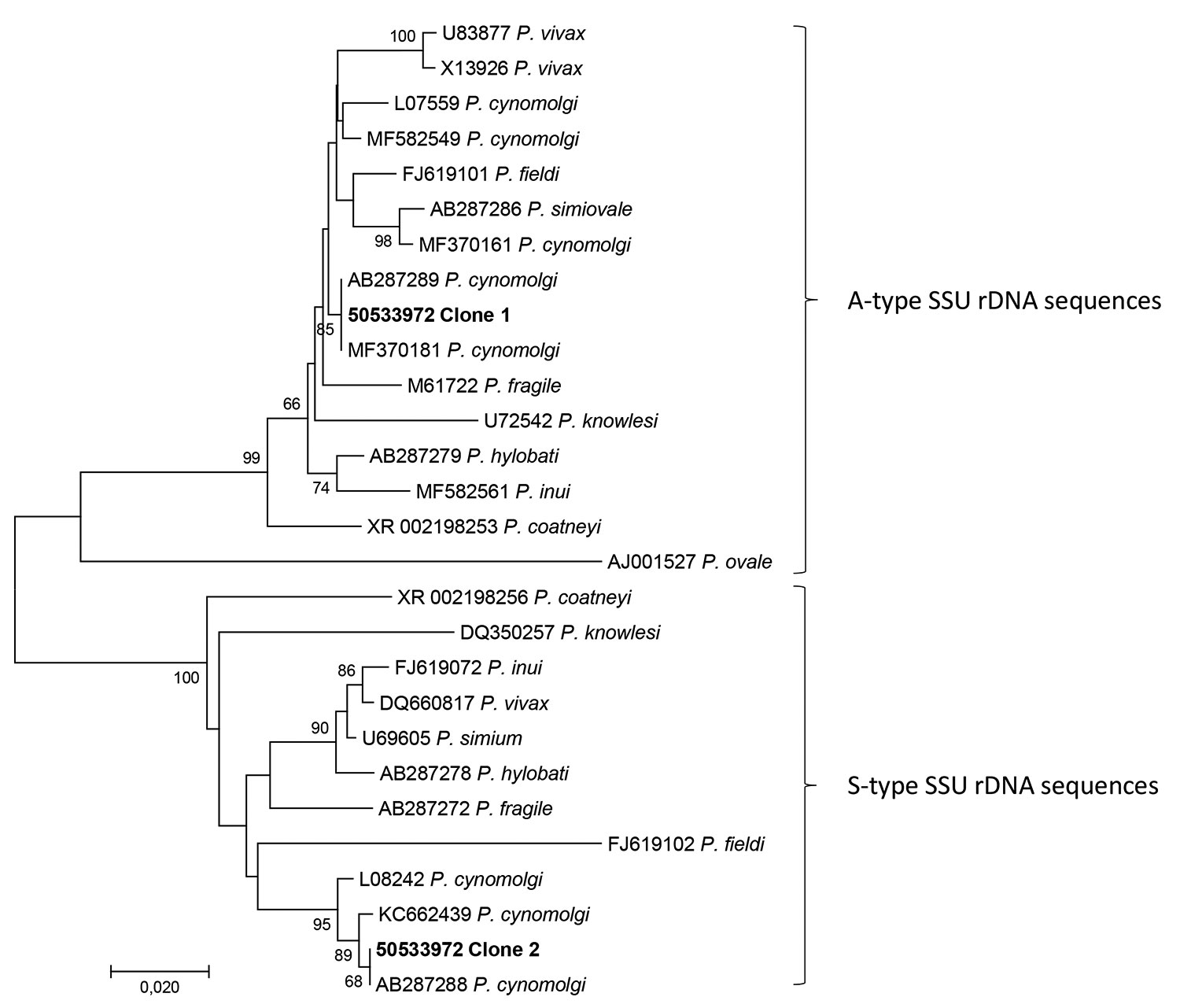
Figure 2. Phylogenetic analysis of the 2 consensus sequences (50533972 clone 1 and 50533972 clone 2) generated by the microbiome assay of blood from a traveler returning from Southeast Asia to Denmark. We used CD-HIT Suite (http://weizhong-lab.ucsd.edu/cdhit_suite/cgi-bin/index.cgi?cmd=cd-hit-est) to cluster sequences reflecting Plasmodium-specific DNA amplified and sequenced by our microbiome assay; we generated consensus sequences using an in-house sequence clustering software. We queried the 2 resulting consensus sequences in GenBank, then downloaded examples of DNA sequences with varying genetic similarity and included them in a multiple sequence alignment with the 2 consensus sequences. Phylogenetic analysis revealed that the microbiome assay had amplified asexual stage-specific (A-type) SSU rRNA genes of Plasmodium cynomolgi, with 50533972 clone 1 reflecting them, and sporozoite stage-specific (S-type), with 50533972 clone 2 reflecting them. We conducted phylogenetic analysis involving 28 DNA sequences in MEGA7 (http:/www.megasoftware.net) and included a total of 464 positions in the final dataset. We inferred evolutionary history using the neighbor-joining method. Numbers at the branches show the percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. We computed evolutionary distances using the Kimura 2-parameter method. Scale bar indicates nucleotide substitutions per site.