Volume 25, Number 10—October 2019
Research Letter
Emergence of Influenza A(H7N4) Virus, Cambodia
Figure
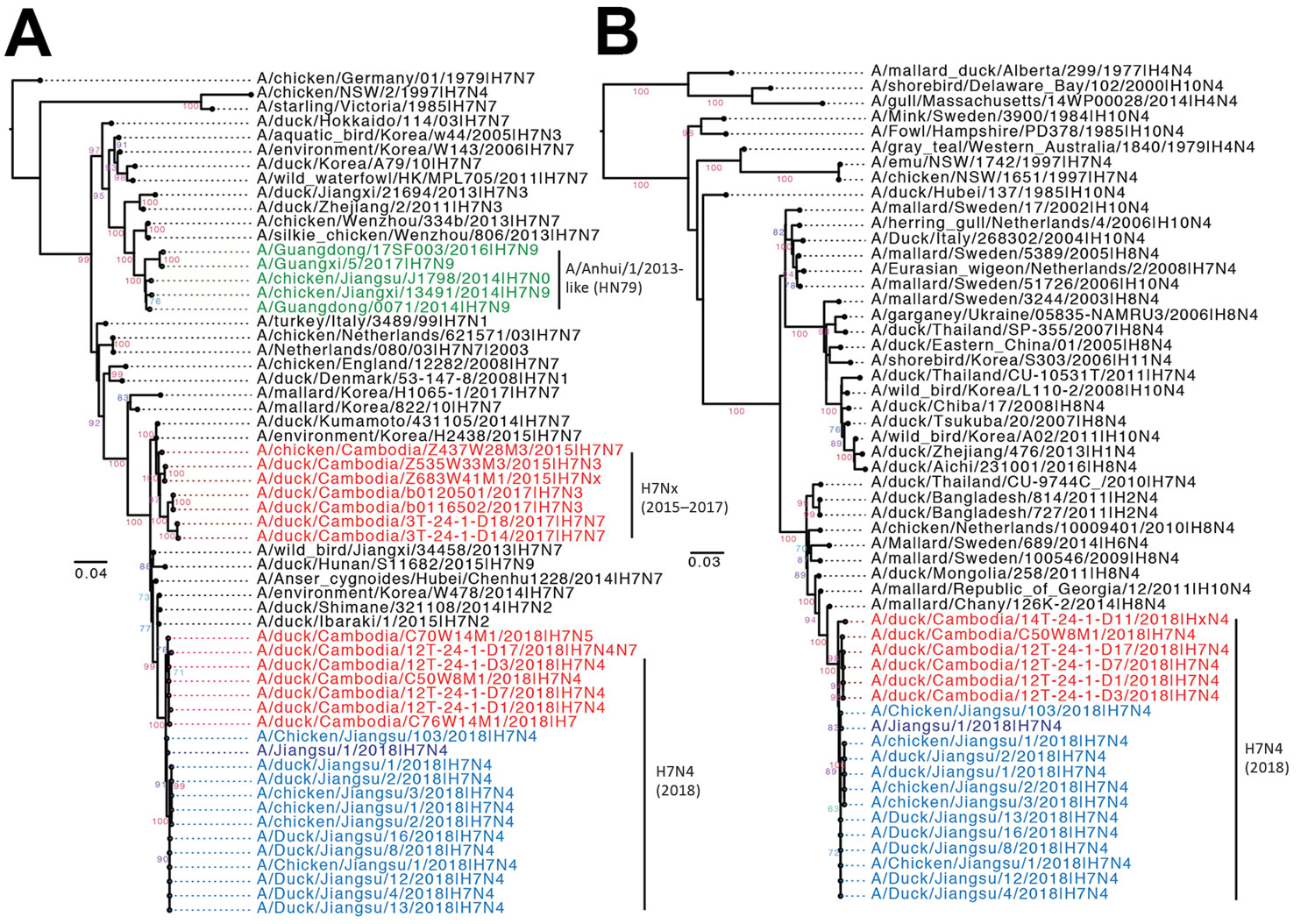
Figure. Maximum-likelihood phylogeny of the evolutionary origins of influenza A(H7N4) virus in Cambodia and comparison with reference isolates. H7 hemagglutinin (A) and N4 neuraminidase (B) genes were inferred using a general time-reversible nucleotide substitution model with a gamma distribution of among-site rate variation in RAxML version 8 (https://cme.h-its.org/exelixis/web/software/raxml) and visualized using Figtree version 1.4 (http://tree.bio.ed.ac.uk/software/figtree/). Branch support values were generated using 1,000 bootstrap replicates. Green indicates A/Anhui 1/2013-like lineage viruses; red indicates viruses from Cambodia; blue indicates A/Jiangsu/2018-like viruses. Scale bars represent nucleotide substitutions per site.
Page created: September 17, 2019
Page updated: September 17, 2019
Page reviewed: September 17, 2019
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.