Volume 25, Number 11—November 2019
Research
Comparison of Whole-Genome Sequences of Legionella pneumophila in Tap Water and in Clinical Strains, Flint, Michigan, USA, 2016
Figure
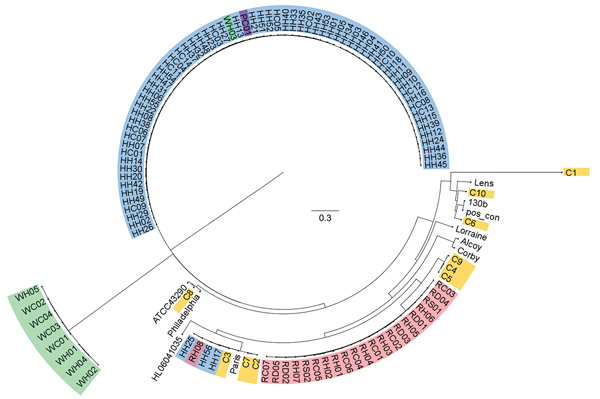
Figure. Single-nucleotide polymorphism (SNP) analysis of isolates from study of Legionella pneumophila in tap water, Flint, Michigan, USA. Analysis was conducted in kSNP3.0 (https://sourceforge.net/projects/ksnp/) and visualized by using FigTree 14.3 (https://github.com/rambaut/figtree/releases/tag/v1.4.3). Isolate sources: yellow, clinical samples; blue, hospital water; red, residence water; purple, public building water; green, buildings supplied by well water. With the exception of buildings supplied by well water, all buildings were serviced by Flint municipal water. Reference strains are detailed in Appendix 1 Table 2. Scale bar indicates nucleotide substitutions per site.
Page created: October 15, 2019
Page updated: October 15, 2019
Page reviewed: October 15, 2019
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.