Volume 25, Number 12—December 2019
Dispatch
Divergent Barmah Forest Virus from Papua New Guinea
Figure 1
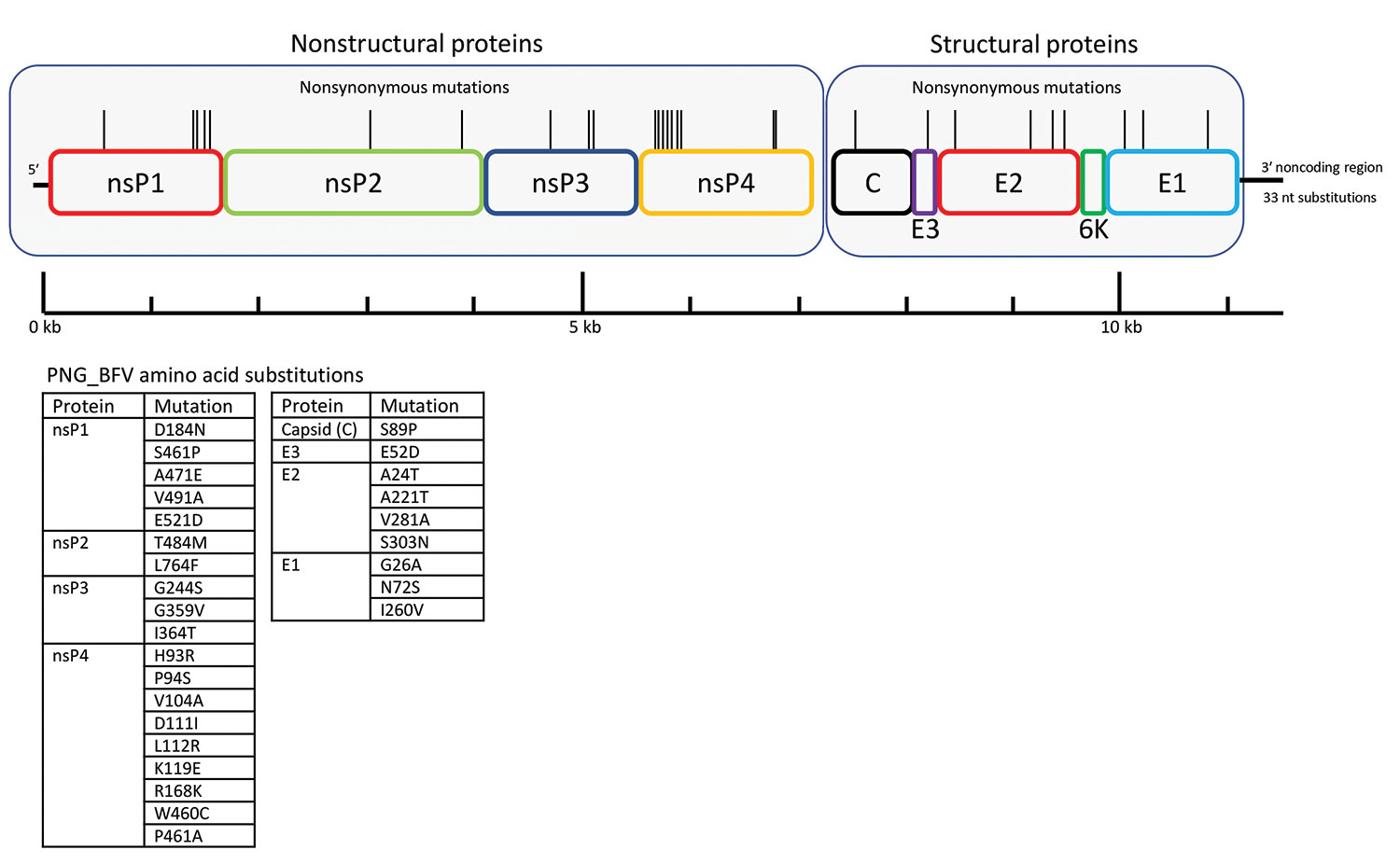
Figure 1. Schematic representation of the BFV genome showing location of amino acid differences between the PNG_BFV (MN115377) isolate from a child in Papua New Guinea and prototype strain BH2193 (RefSeq accession no. NC_001786.1). Amino acid substitutions in the PNG_BFV genome are shown in nonstructural proteins nsP1–4 (n = 19) and structural proteins C, E1–3, and 6K (n = 9) and listed below the schematic. BFV, Barmah Forest virus; C, capsid; E, envelope; nsP, nonstructural protein; PNG, Papua New Guinea.
Page created: November 18, 2019
Page updated: November 18, 2019
Page reviewed: November 18, 2019
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.